SLO co-opts host cell glycosphingolipids to access cholesterol-rich lipid rafts for enhanced pore formation and cytotoxicity
- PMID: 39835825
- PMCID: PMC11898750
- DOI: 10.1128/mbio.03777-24
SLO co-opts host cell glycosphingolipids to access cholesterol-rich lipid rafts for enhanced pore formation and cytotoxicity
Abstract
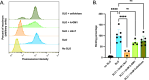
Streptolysin O (SLO) is a virulence determinant of group A Streptococcus (S. pyogenes), the agent of streptococcal sore throat and severe invasive infections. SLO is a member of a family of bacterial pore-forming toxins known as cholesterol-dependent cytolysins, which require cell membrane cholesterol for pore formation. While cholesterol is essential for cytolytic activity, accumulating data suggest that cell surface glycans may also participate in the binding of SLO and other cholesterol-dependent cytolysins to host cells. Here, we find that unbiased CRISPR screens for host susceptibility factors for SLO cytotoxicity identified genes encoding enzymes involved in the earliest steps of glycosphingolipid (GSL) biosynthesis. Targeted knockouts of these genes conferred relative resistance to SLO cytotoxicity in two independent human cell lines. Inactivation of ugcg, which codes for UDP-glucose ceramide glucosyltransferase, the enzyme catalyzing the first glycosylation step in GSL biosynthesis, reduced the clustering of SLO on the cell surface. This result suggests that binding to GSLs serves to cluster SLO molecules at lipid rafts where both GSLs and cholesterol are abundant. SLO clustering and susceptibility to SLO cytotoxicity were restored by reconstituting the GSL content of ugcg knockout cells with ganglioside GM1, but susceptibility to SLO cytotoxicity was not restored by a GM1 variant that lacks an oligosaccharide head group required for SLO binding, nor by a variant with a "kinked" acyl chain that prevents efficient packing of the ganglioside ceramide moiety with cholesterol. Thus, SLO appears to co-opt cell surface glycosphingolipids to gain access to lipid rafts for increased efficiency of pore formation and cytotoxicity.
Importance: Group A Streptococcus is a global public health concern as it causes streptococcal sore throat and less common but potentially life-threatening invasive infections. Invasive infections have been associated with bacterial strains that produce large amounts of a secreted toxin, streptolysin O (SLO), which belongs to a family of pore-forming toxins produced by a variety of bacterial species. This study reveals that SLO binds to a class of molecules known as glycosphingolipids on the surface of human cells and that this interaction promotes efficient binding of SLO to cholesterol in the cell membrane and enhances pore formation. Understanding how SLO damages human cells provides new insight into streptococcal infection and may inform new approaches to treatment and prevention.
Keywords: Streptococcus pyogenes; cholesterol-dependent cytolysin; glycan; glycosphingolipid; group A Streptococcus; lipid raft; receptor; streptolysin O; toxin.
Conflict of interest statement
W.I.L. is a scientific founder and board member of Transcera Inc. The company is developing the use of modified ceramides as vehicles for drug delivery and lysosomal degradation of pathogenic proteins. All other authors declare no competing interests.
Figures





References
-
- Harder J, Franchi L, Muñoz-Planillo R, Park J-H, Reimer T, Núñez G. 2009. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-kappa B activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol 183:5823–5829. doi:10.4049/jimmunol.0900444 - DOI - PMC - PubMed
-
- Malley R, Henneke P, Morse SC, Cieslewicz MJ, Lipsitch M, Thompson CM, Kurt-Jones E, Paton JC, Wessels MR, Golenbock DT. 2003. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci U S A 100:1966–1971. doi:10.1073/pnas.0435928100 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- AI112823/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- AI130019/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- DK 048106, DK122953, DK 034854/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK048106/DK/NIDDK NIH HHS/United States
- R01 AI130019/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
