The PurR family transcriptional regulator promotes butenyl-spinosyn production in Saccharopolyspora pogona
- PMID: 39836216
- PMCID: PMC11750948
- DOI: 10.1007/s00253-024-13390-1
The PurR family transcriptional regulator promotes butenyl-spinosyn production in Saccharopolyspora pogona
Abstract
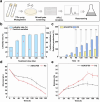
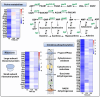
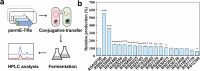
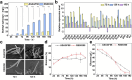
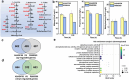
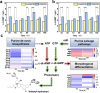
Butenyl-spinosyn, derived from Saccharopolyspora pogona, is a broad-spectrum and effective bioinsecticide. However, the regulatory mechanism affecting butenyl-spinosyn synthesis has not been fully elucidated, which hindered the improvement of production. Here, a high-production strain S. pogona H2 was generated by Cobalt-60 γ-ray mutagenesis, which showed a 2.7-fold increase in production compared to the wild-type strain S. pogona ASAGF58. A comparative transcriptomic analysis between S. pogona ASAGF58 and H2 was performed to elucidate the high-production mechanism that more precursors and energy were used to synthesize of butenyl-spinosyn. Fortunately, a PurR family transcriptional regulator TF00350 was discovered. TF00350 overexpression strain RS00350 induced morphological differentiation and butenyl-spinosyn production, ultimately leading to a 5.5-fold increase in butenyl-spinosyn production (141.5 ± 1.03 mg/L). Through transcriptomics analysis, most genes related to purine metabolism pathway were downregulated, and the butenyl-spinosyn biosynthesis gene was upregulated by increasing the concentration of c-di-GMP and decreasing the concentration of c-di-AMP. These results provide valuable insights for further mining key regulators and improving butenyl-spinosyn production. KEY POINTS: • A high production strain of S. pogona H2 was obtained by 60Co γ-ray mutagenesis. • Positive regulator TF00350 identified by transcriptomics, increasing butenyl-spinosyn production by 5.5-fold. • TF00350 regulated of butenyl-spinosyn production by second messengers.
Keywords: Saccharopolyspora pogona; Butenyl-spinosyn; PurR family transcriptional regulator; Transcriptomic.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: This article does not contain any studies with human participants performed by any of the authors. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Bacterioferritin: a key iron storage modulator that affects strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Microb Cell Fact. 2021 Aug 14;20(1):157. doi: 10.1186/s12934-021-01651-x. Microb Cell Fact. 2021. PMID: 34391414 Free PMC article.
-
Effect of pII key nitrogen regulatory gene on strain growth and butenyl-spinosyn biosynthesis in Saccharopolyspora pogona.Appl Microbiol Biotechnol. 2022 Apr;106(8):3081-3091. doi: 10.1007/s00253-022-11902-5. Epub 2022 Apr 4. Appl Microbiol Biotechnol. 2022. PMID: 35376972
-
Comparative Proteomics Reveals the Effect of the Transcriptional Regulator Sp13016 on Butenyl-Spinosyn Biosynthesis in Saccharopolyspora pogona.J Agric Food Chem. 2021 Oct 27;69(42):12554-12565. doi: 10.1021/acs.jafc.1c03654. Epub 2021 Oct 16. J Agric Food Chem. 2021. PMID: 34657420
-
Recent advances in the biochemistry of spinosyns.Appl Microbiol Biotechnol. 2009 Feb;82(1):13-23. doi: 10.1007/s00253-008-1784-8. Epub 2008 Dec 10. Appl Microbiol Biotechnol. 2009. PMID: 19082588 Review.
-
Genes for the biosynthesis of spinosyns: applications for yield improvement in Saccharopolyspora spinosa.J Ind Microbiol Biotechnol. 2001 Dec;27(6):399-402. doi: 10.1038/sj.jim.7000180. J Ind Microbiol Biotechnol. 2001. PMID: 11774006 Review.
Cited by
-
Ribosome Engineering for Enhanced Butenyl-Spinosyn Production in Saccharopolyspora pogona.Curr Microbiol. 2025 Jun 17;82(8):337. doi: 10.1007/s00284-025-04317-8. Curr Microbiol. 2025. PMID: 40526196
References
-
- Alvarez AF, Georgellis D (2023) Environmental adaptation and diversification of bacterial two-component systems. Curr Opin Microbiol 76:102399. 10.1016/j.mib.2023.102399 - PubMed
-
- Anderson BW, Schumacher MA, Yang J, Turdiev A, Turdiev H, Schroeder JW, He Q, Lee VT, Brennan RG, Wang JD (2022) The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus. Nucleic Acids Res 50(2):847–866. 10.1093/nar/gkab1281 - PMC - PubMed
-
- Bierman M, Logan R, O’Brien K, Seno ET, Nagaraja RR, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49. 10.1016/0378-1119(92)90627-2 - PubMed
-
- Bignell DRD, Francis IM, Fyans JK, Loria R (2014) Thaxtomin A production and virulence are controlled by several bld gene global regulators in Streptomyces scabies. Mol Plant Microbe in 27(8):875–885. 10.1094/MPMI-02-14-0037-R - PubMed
-
- Cai X, Jin JY, Zhang B, Liu ZQ, Zheng YG (2021) Improvement of cordycepin production by an isolated Paecilomyces hepiali mutant from combinatorial mutation breeding and medium screening. Bioprocess Biosystems Eng 44(11):2387–2398. 10.1007/s00449-021-02611-w - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

