NOL-7 serves as a potential prognostic-related biomarker for hepatocellular carcinoma
- PMID: 39836310
- PMCID: PMC11751243
- DOI: 10.1007/s12672-024-01551-7
NOL-7 serves as a potential prognostic-related biomarker for hepatocellular carcinoma
Abstract
Background: Nucleolar protein 7 (NOL7), a specific protein found in the nucleolus, is crucial for maintaining cell division and proliferation. While the involvement of NOL7 in influencing the unfavorable prognosis of metastatic melanoma has been reported, its significance in predicting the prognosis of patients with Hepatocellular Carcinoma (HCC) remains unclear.
Methods: Aberrant expression of NOL7 in HCC and its prognostic value were evaluated using multiple databases, including TCGA, GTEx, Xiantao Academic, HCCDB, UALCAN, TISCH, and STRING. Immunohistochemistry (IHC) and quantitative real-time PCR were used to validate NOL7 expression levels in patients with HCC.
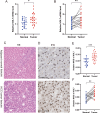
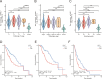
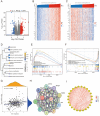
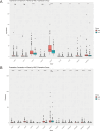
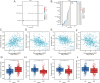
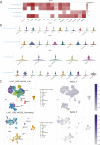
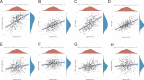
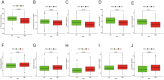
Results: NOL7 expression was higher in the HCC samples than in the normal samples (P < 0.05). NOL7 was strongly associated with elevated AFP levels, vascular invasion, TNM stage, poorer tumor differentiation, and poorer survival (all P < 0.05). Elevated NOL7 expression correlated with decreased overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) (all P < 0.05). Multivariate analysis revealed that NOL7 was an independent prognostic factor that was significantly related to OS and DSS. The nomogram showed a good predictive performance based on the calibration plot. In addition, NOL7 expression was significantly correlated with cell cycle modulators, immune checkpoints, and various immune cell populations. In addition, we identified eight potential pathways associated with NOL7 as the most promising pathways for NOL7 in HCC. Low-risk specimens were more sensitive to oxaliplatin, cisplatin, irinotecan, sorafenib, and cytarabine than high-risk specimens.
Conclusion: NOL7 may serve as a potential biomarker for predicting clinical outcomes and may provide guidance for clinical therapy in patients with HCC.
Keywords: Biomarker; Chemosensitivity; Hepatocellular carcinoma; NOL7; Tumor immune microenvironment.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was carried out in line with the principles of the Declaration of Helsinki. The protocol was approved by the institutional review board of The First People’s Hospital of Foshan (approval number: medical ethics 2023 No. 74). All methods have been performed in accordance with the regulations of the Institutional Research Ethics Committee at Foshan First People’ s Hospital. All patients or their authorized family members in the study provided thorough and accurate informed consent prior to inclusion. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Cell division cycle-associated 8 is a prognostic biomarker related to immune invasion in hepatocellular carcinoma.Cancer Med. 2023 Apr;12(8):10138-10155. doi: 10.1002/cam4.5718. Epub 2023 Feb 28. Cancer Med. 2023. PMID: 36855818 Free PMC article.
-
Study of TRAF3IP3 for prognosis and immune infiltration in hepatocellular carcinoma.PeerJ. 2024 Dec 12;12:e18538. doi: 10.7717/peerj.18538. eCollection 2024. PeerJ. 2024. PMID: 39677949 Free PMC article.
-
CENPO is Associated with Immune Cell Infiltration and is a Potential Diagnostic and Prognostic Marker for Hepatocellular Carcinoma.Int J Gen Med. 2022 Sep 25;15:7493-7510. doi: 10.2147/IJGM.S382234. eCollection 2022. Int J Gen Med. 2022. PMID: 36187159 Free PMC article.
-
INKA2-AS1 Is a Potential Promising Prognostic-Related Biomarker and Correlated with Immune Infiltrates in Hepatocellular Carcinoma.Mediators Inflamm. 2023 May 2;2023:7057236. doi: 10.1155/2023/7057236. eCollection 2023. Mediators Inflamm. 2023. PMID: 37181806 Free PMC article.
-
Pancancer Analysis of the Oncogenic and Prognostic Role of NOL7: A Potential Target for Carcinogenesis and Survival.Int J Mol Sci. 2022 Aug 25;23(17):9611. doi: 10.3390/ijms23179611. Int J Mol Sci. 2022. PMID: 36077008 Free PMC article.
References
-
- Ganesan P, Kulik LM. Hepatocellular carcinoma: new developments. Clin Liver Dis. 2023;27(1):85–102. 10.1016/j.cld.2022.08.004. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. - PubMed
-
- Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated proteogenomic characterization of hbv-related hepatocellular carcinoma. Cell. 2019;179(2):561-577.e22. 10.1016/j.cell.2019.08.052. - PubMed
-
- Allaire M, Nault JC. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29(4):288–95. 10.1097/CCO.0000000000000378. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
