Mycobacterium tuberculosis resisters despite HIV exhibit activated T cells and macrophages in their pulmonary alveoli
- PMID: 39836471
- PMCID: PMC11957701
- DOI: 10.1172/JCI188016
Mycobacterium tuberculosis resisters despite HIV exhibit activated T cells and macrophages in their pulmonary alveoli
Abstract
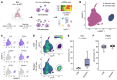
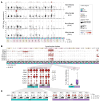
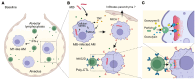
BACKGROUNDNatural resistance to Mycobacterium tuberculosis (Mtb) infection in some people with HIV (PWH) is unexplained.METHODSWe performed single cell RNA-sequencing of bronchoalveolar lavage cells, unstimulated or ex vivo stimulated with Mtb, for 7 PWH who were tuberculin skin test (TST) and IFN-γ release assay (IGRA) positive (called LTBI) and 6 who were persistently TST and IGRA negative (called resisters).RESULTSAlveolar macrophages (AM) from resisters displayed a baseline M1 macrophage phenotype while AM from LTBI did not. Resisters displayed alveolar lymphocytosis, with enrichment of all T cell subpopulations including IFNG-expressing cells. In both groups, mycobactericidal granulysin was expressed almost exclusively by a T cell subtype that coexpressed granzyme B, perforin and NK cell receptors. These poly-cytotoxic T lymphocytes (poly-CTL) overexpressed activating NK cell receptors and were increased in resister BAL. Following challenge with Mtb, only intraepithelial lymphocyte-like cells from LTBI participants responded with increased transcription of IFNG. AM from resisters responded with a stronger TNF signature at 6 hours after infection while at 24 hours after infection, AM from LTBI displayed a stronger IFN-γ signature. Conversely, at 24 hours after infection, only AM from resisters displayed an upregulation of MHC class I polypeptide-related sequence A (MICA) transcripts, which encode an activating ligand for poly-CTL.CONCLUSIONThese results suggest that poly-CTL and M1-like pre-activated AM mediate the resister phenotype in PWH.FUNDINGNational Institutes of Health. Canadian Institutes of Health Research. Digital Research Alliance of Canada. French National Research Agency. French National Agency for Research on AIDS and Viral Hepatitis. St. Giles Foundation. General Atlantic Foundation. South African Medical Research Council Centre for Tuberculosis Research.
Keywords: Infectious disease; Macrophages; T cells; Tuberculosis.
Figures




Update of
-
Mycobacterium tuberculosis resisters despite HIV exhibit activated T cells and macrophages in their pulmonary alveoli.Res Sq [Preprint]. 2024 Jan 31:rs.3.rs-3889020. doi: 10.21203/rs.3.rs-3889020/v1. Res Sq. 2024. Update in: J Clin Invest. 2025 Jan 21;135(7):e188016. doi: 10.1172/JCI188016. PMID: 38352496 Free PMC article. Updated. Preprint.
References
-
- WHO. Global Tuberculosis Report 2023. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... Accessed January 22, 2025.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

