The impact of remdesivir on SARS-CoV-2 evolution in vivo
- PMID: 39836474
- PMCID: PMC11949014
- DOI: 10.1172/jci.insight.182376
The impact of remdesivir on SARS-CoV-2 evolution in vivo
Abstract
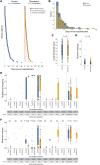
The impact of remdesivir on SARS-CoV-2 diversity and evolution in vivo has remained unclear. In this single-center, retrospective cohort study, we assessed SARS-CoV-2 diversification and diversity over time in a cohort of hospitalized patients who did or did not receive remdesivir. Whole-genome sequencing was performed on 98 paired specimens collected from 49 patients before and after remdesivir administration. The genetic divergence between paired specimens was not significantly different in this cohort compared with that in a control group of patients who did not receive the drug. However, when we focused on minority variants, several positions showed preferential diversification after remdesivir treatment, some of which were associated with specific variants of concern. Most notably, remdesivir administration resulted in strong selection for a nonsynonymous mutation in nsp12, G671S, previously associated with enhanced viral fitness. This same mutation was found to be enriched in a second cohort of 143 inpatients with specimens collected after remdesivir administration compared with controls. Only one other mutation previously implicated in remdesivir resistance (nsp12:V792I) was found to be preferentially selected for after remdesivir administration. These data suggest that SARS-CoV-2 variants with enhanced replicative fitness may be selected for in the presence of antiviral therapy as an indirect means to overcome this selective pressure.
Keywords: COVID-19; Drug therapy; Therapeutics.
Conflict of interest statement
Figures




References
-
- WHO Coronavirus (COVID-19) Dashboard. https://data.who.int/dashboards/covid19/cases Updated December 29, 2024. Accessed January 15, 2025.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

