The levels of the long noncoding RNA MALAT1 affect cell viability and modulate TDP-43 binding to mRNA in the nucleus
- PMID: 39837396
- PMCID: PMC11871449
- DOI: 10.1016/j.jbc.2025.108207
The levels of the long noncoding RNA MALAT1 affect cell viability and modulate TDP-43 binding to mRNA in the nucleus
Abstract
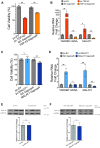
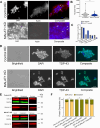
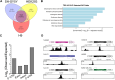
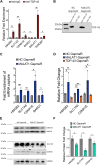
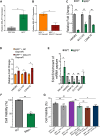
TAR DNA-binding protein (TDP-43) and metastasis-associated lung adenocarcinoma transcript (MALAT1) RNA are both abundantly expressed in the human cell nucleus. Increased interaction of TDP-43 and MALAT1, as well as dysregulation of TDP-43 function, was previously identified in brain samples from patients with neurodegenerative disease compared to healthy brain tissues. We hypothesized that TDP-43 function may depend in part on MALAT1 expression levels. Here, we find that alterations in MALAT1 expression affect cell viability and can modulate TDP-43 binding to other mRNAs in HEK293 and SH-SY5Y human cell lines. Disruption of either MALAT1 or TDP-43 expression induces cell death, indicating that both macromolecules contribute positively to survival. Depletion of MALAT1 RNA results in increased binding of TDP-43 to other mRNA transcripts at the 3' UTR. Finally, we examined the contribution of MALAT1 expression to survival in a cell culture model of neurodegeneration using MPP+ treatment in SH-SY5Y cells. Depletion of MALAT1 RNA protects against toxicity in a cellular model of neurodegeneration and modulates TDP-43 binding to mRNA transcripts involved in apoptotic cell death. Taken together, we find that MALAT1 RNA and TDP-43 interactions can affect mRNA levels and cell viability. A tightly regulated network of noncoding RNA, messenger RNA, and protein interactions could provide a mechanism to maintain appropriate RNA expression levels and contribute to neuronal function.
Keywords: Long non-coding RNA; MALAT1; RNA binding protein; RNA-protein interaction; TAR DNA-binding protein 43 (TDP-43) (TARDBP); gene expression; mRNA; mRNA stability; neurodegeneration.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. - PubMed
-
- Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
-
- Hasegawa M., Arai T., Akiyama H., Nonaka T., Mori H., Hashimoto T., et al. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain. 2007;130:1386–1394. - PubMed
-
- Nakashima-Yasuda H., Uryu K., Robinson J., Xie S.X., Hurtig H., Duda J.E., et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. - PubMed

