Barcoded screening identifies nanocarriers for protein delivery to kidney
- PMID: 39837887
- PMCID: PMC11751284
- DOI: 10.1038/s41467-025-56257-3
Barcoded screening identifies nanocarriers for protein delivery to kidney
Abstract
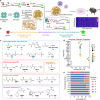
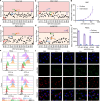
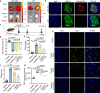
Targeted protein delivery with nanocarriers holds significant potential to enhance therapeutic outcomes by precisely directing proteins to specific organs or tissues. However, the complex interactions between nanocarriers and the biological environment pose considerable challenges in designing effective targeted delivery vehicles. In this study, we address this challenge by leveraging DNA-barcoded high-throughput screening. We construct a nanocapsule library via in-situ polymerization, incorporating various monomers to create nanocapsules with unique surface properties. In vitro and in vivo screening, using female mice, identify nanocapsules with high cell association and different biodistribution. Our investigation into kidney-enriched nanocapsules highlights the crucial role of polymer composition in biodistribution, demonstrating the potential of surface engineering for precise control over nanoparticle distribution. The kidney-enriched nanocapsule successfully delivers catalase, showcasing its therapeutic potential in mitigating cisplatin-induced acute kidney injury. Overall, our study presents an approach for identifying protein delivery vehicles, with the capacity to broaden the application of proteins as therapeutic agents or research tools.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interest.
Figures



Similar articles
-
Improved bioavailability of polydatin and its protective effect against cisplatin induced nephrotoxicity through self-assembled fucoidan and carboxymethyl chitosan delivery system.Int J Biol Macromol. 2025 Jan;287:138577. doi: 10.1016/j.ijbiomac.2024.138577. Epub 2024 Dec 9. Int J Biol Macromol. 2025. PMID: 39657878
-
Characterization of different functionalized lipidic nanocapsules as potential drug carriers.Int J Mol Sci. 2012;13(2):2405-2424. doi: 10.3390/ijms13022405. Epub 2012 Feb 22. Int J Mol Sci. 2012. PMID: 22408461 Free PMC article.
-
Redox-responsive nanocapsules for intracellular protein delivery.Biomaterials. 2011 Aug;32(22):5223-30. doi: 10.1016/j.biomaterials.2011.03.060. Epub 2011 Apr 22. Biomaterials. 2011. PMID: 21514660
-
Lipid Nanocapsule: A Novel Approach to Drug Delivery System Formulation Development.Curr Pharm Biotechnol. 2024;25(3):268-284. doi: 10.2174/1389201024666230523114350. Curr Pharm Biotechnol. 2024. PMID: 37231750 Review.
-
Intracellular delivery of proteins by nanocarriers.Nanomedicine (Lond). 2017 Apr;12(8):941-952. doi: 10.2217/nnm-2016-0393. Epub 2017 Mar 24. Nanomedicine (Lond). 2017. PMID: 28338410 Free PMC article. Review.
Cited by
-
Targeting ion channel networks in diabetic kidney disease: from molecular crosstalk to precision therapeutics and clinical innovation.Front Med (Lausanne). 2025 Jun 26;12:1607701. doi: 10.3389/fmed.2025.1607701. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40641971 Free PMC article. Review.
References
-
- Kim, J., Eygeris, Y., Ryals, R. C., Jozic, A. & Sahay, G. Strategies for non-viral vectors targeting organs beyond the liver. Nat. Nanotechnol.19, 428–447 (2023). - PubMed
-
- Su, H., Rong, G., Li, L. & Cheng, Y. Subcellular targeting strategies for protein and peptide delivery. Adv. Drug. Deliver. Rev. 212, 10.1016/j.addr.2024.115387 (2024). - PubMed
-
- Cabral, H., Li, J., Miyata, K. & Kataoka, K. Controlling the biodistribution and clearance of nanomedicines. Nat. Rev. Bioeng.2, 214–232 (2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

