Targeting BRCA1-deficient PARP inhibitor-resistant cells with nickases reveals nick resection as a cancer vulnerability
- PMID: 39838098
- PMCID: PMC12041741
- DOI: 10.1038/s43018-024-00902-1
Targeting BRCA1-deficient PARP inhibitor-resistant cells with nickases reveals nick resection as a cancer vulnerability
Abstract
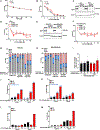
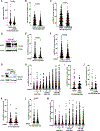
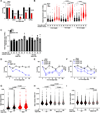
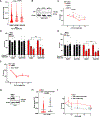
Tumors lacking the BRCA1 and BRCA2 (BRCA) hereditary breast cancer genes display heightened sensitivity to anti-cancer treatments, such as inhibitors of poly (ADP-ribose) polymerase 1 (PARP1). However, when resistance develops, treatments are lacking. Using CRISPR technology, we discovered that enhancing homologous recombination through increased DNA end resection in BRCA1-deficient cells by loss of the 53BP1-Shieldin complex-which is associated with resistance to PARP inhibitors-also heightens sensitivity to DNA nicks. The sensitivity is caused by hyper-resection of nicks into extensive single-stranded regions that trigger cell death. Based on these findings and that nicks limit tumor formation in mice, we propose nickases as a tool for personalized medicine. Moreover, our findings indicate that restricting nick expansion is a critical function of the 53BP1-Shieldin complex.
© 2025. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: Authors declare no competing interests. Inclusion & ethics statement: Each collaborator in this study has satisfied the authorship criteria set by Nature Portfolio journals, earning inclusion as an author. Their contributions to planning and performing the study were appropriately acknowledged. Collaborators established roles and responsibilities prior to initiating the research.
Figures







References
-
- Futreal PA et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266, 120–122 (1994). - PubMed
-
- Bryant HE et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). - PubMed
-
- Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). - PubMed
MeSH terms
Substances
Grants and funding
- R01CA285607/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- 1F32CA268524-01A1/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- R01 CA285607/CA/NCI NIH HHS/United States
- F32 CA077880/CA/NCI NIH HHS/United States
- R01 CA254037/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

