The Shu complex interacts with the replicative helicase to prevent mutations and aberrant recombination
- PMID: 39838174
- PMCID: PMC11876325
- DOI: 10.1038/s44318-025-00365-9
The Shu complex interacts with the replicative helicase to prevent mutations and aberrant recombination
Abstract
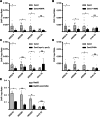
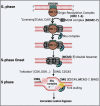
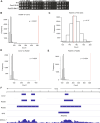
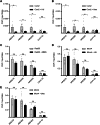
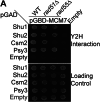
Homologous recombination (HR) is important for DNA damage tolerance during replication. The yeast Shu complex, a conserved homologous recombination factor, prevents replication-associated mutagenesis. Here we examine how yeast cells require the Shu complex for coping with MMS-induced lesions during DNA replication. We find that Csm2, a subunit of the Shu complex, binds to autonomous-replicating sequences (ARS) in yeast. Further evolutionary studies reveal that the yeast and human Shu complexes have co-evolved with specific replication-initiation factors. The connection between the Shu complex and replication is underlined by the finding that the Shu complex interacts with the ORC and MCM complexes. For example, the Shu complex interacts, independent of other HR proteins, with the replication initiation complexes through the N-terminus of Psy3. Lastly, we show interactions between the Shu complex and the replication initiation complexes are essential for resistance to DNA damage, to prevent mutations and aberrant recombination events. In our model, the Shu complex interacts with the replication machinery to enable error-free bypass of DNA damage.
Keywords: DNA Damage Tolerance; DNA Replication; Homologous Recombination; RAD51; Shu Complex.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures







References
-
- Ball LG, Zhang K, Cobb JA, Boone C, Xiao W (2009) The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol Microbiol 73:89–102 - PubMed
-
- Bauerschmidt C, Pollok S, Kremmer E, Nasheuer H-P, Grosse F (2007) Interactions of human Cdc45 with the Mcm2-7 complex, the GINS complex, and DNA polymerases delta and epsilon during S phase. Genes Cells 12:745–758 - PubMed
MeSH terms
Substances
Grants and funding
- BONFOR 2020-1B-03/Uni Bonn | Medizinische Fakultät, Rheinische Friedrich-Wilhelms-Universität Bonn (Faculty of Medicine, Uni Bonn)
- P30 ES013508/ES/NIEHS NIH HHS/United States
- EXC2151 - 390873048/Deutsche Forschungsgemeinschaft (DFG)
- R01 HG009299/HG/NHGRI NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

