Oncogenic mutant KRAS inhibition through oxidation at cysteine 118
- PMID: 39838816
- PMCID: PMC11793020
- DOI: 10.1002/1878-0261.13798
Oncogenic mutant KRAS inhibition through oxidation at cysteine 118
Abstract
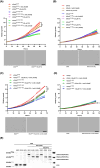
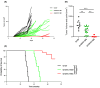
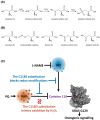
Specific reactive oxygen species activate the GTPase Kirsten rat sarcoma virus (KRAS) by reacting with cysteine 118 (C118), leading to an electron transfer between C118 and nucleoside guanosine diphosphate (GDP), which causes the release of GDP. Here, we have mimicked permanent oxidation of human KRAS at C118 by replacing C118 with aspartic acid (C118D) in KRAS to show that oncogenic mutant KRAS is selectively inhibited via oxidation at C118, both in vitro and in vivo. Moreover, the combined treatment of hydrogen-peroxide-producing pro-oxidant paraquat and nitric-oxide-producing inhibitor N(ω)-nitro-l-arginine methyl ester selectively inhibits human mutant KRAS activity by inducing oxidization at C118. Our study shows for the first time the vulnerability of human mutant KRAS to oxidation, thereby paving the way to explore oxidation-based anti-KRAS treatments in humans.
Keywords: KRAS C118; NSCLC; ROS; cysteine modification; oncogene; redox regulation.
© 2025 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

