Viral diversity in wild rodents in the regions of Canaã de Carajás and Curionopólis, State of Pará, Brazil
- PMID: 39839123
- PMCID: PMC11747277
- DOI: 10.3389/fmicb.2024.1502462
Viral diversity in wild rodents in the regions of Canaã de Carajás and Curionopólis, State of Pará, Brazil
Abstract
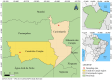
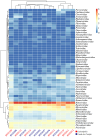
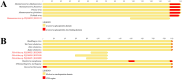
Wild rodents serve as crucial reservoirs for zoonotic viruses. Anthropogenic and environmental disruptions, particularly those induced by mining activities, can destabilize rodent populations and facilitate the emergence of viral agents. In the Canaã dos Carajás and Curionópolis regions of Brazil, significant environmental changes have occurred due to mining expansion, potentially creating conditions conducive to the emergence of rodent-associated viral diseases. This study aimed to investigate the viral diversity in wild rodents captured in Canaã dos Carajás and Curionópolis, Pará, between 2017 and 2019. A total of 102 rodent samples were taxonomically identified through karyotyping and screened for anti-Orthohantavirus antibodies using the ELISA method. Subsequently, nucleotide sequencing and bioinformatics analyses were conducted on 14 selected samples to characterize the virome. This selection was based on the most commonly associated rodent genera as reservoirs of Orthohantavirus and Mammarenavirus. Of the 102 samples tested via ELISA, 100 were negative, and two showed optical density at the cutoff point. Sequencing of the 14 samples generated approximately 520 million reads, with 409 million retained after quality control. These reads were categorized into 53 viral families, including both DNA and RNA viruses, with Retroviridae, Baculoviridae, and Microviridae being the most abundant. Viral contigs were identified, including one fragment related to Arenaviridae and three to Filoviridae. Metagenomic analysis revealed high viral diversity in the sampled rodents, with the presence of viral families of public health concern, such as Arenaviridae and Filoviridae. The findings suggest that increased human activities associated with mining may contribute to the emergence of these viruses, underscoring the need for ongoing surveillance to prevent potential outbreaks.
Keywords: Arenaviridae; Filoviridae; epidemiologic vigilance; metagenomics; rodent.
Copyright © 2025 Monteiro, Silva, Cruz, Silva, Queiroz, Casseb, Martins and Medeiros.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Biedenkopf N., Bukreyev A., Chandran K., Di Paola N., Formenty P. B. H., Griffiths A., et al. . (2023). Renaming of genera Ebolavirus and Marburgvirus to Orthoebolavirus and Orthomarburgvirus, respectively, and introduction of binomial species names within family Filoviridae. Arch. Virol. 168:220. 10.1007/s00705-023-05834-2 - DOI - PubMed
-
- Bovincino C. R., Oliveira J. A., Andrea P. S. (2008). Guia dos roedores do Brasil, com chave para gêneros baseadas em caracteres externos. Rio de Janeiro: Centro Pan–Americano de Febre Aftosa, OPAS/OMS, 122.
LinkOut - more resources
Full Text Sources

