NR2F6 regulates stem cell hematopoiesis and myelopoiesis in mice
- PMID: 39840064
- PMCID: PMC11747239
- DOI: 10.3389/fimmu.2024.1404805
NR2F6 regulates stem cell hematopoiesis and myelopoiesis in mice
Abstract
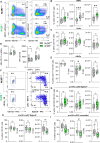
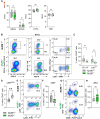
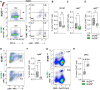
Nuclear receptors regulate hematopoietic stem cells (HSCs) and peripheral immune cells in mice and humans. The nuclear orphan receptor NR2F6 (EAR-2) has been shown to control murine hematopoiesis. Still, detailed analysis of the distinct stem cell, myeloid, and lymphoid progenitors in the bone marrow in a genetic loss of function model remains pending. In this study, we found that adult germline Nr2f6-deficient mice contained increased percentages of total long-term and short-term HSCs, as well as a subpopulation within the lineage-biased multipotent progenitor (MPP3) cells. The loss of NR2F6 thus led to an increase in the percentage of LSK+ cells. Following the differentiation from the common myeloid progenitors (CMP), the granulocyte-monocyte progenitors (GMP) were decreased, while monocyte-dendritic progenitors (MDP) were increased in Nr2f6-deficient bone marrow. Within the pre-conventional dendritic progenitors (pre-cDCs), the subpopulation of pre-cDC2s was reduced in the bone marrow of Nr2f6-deficient mice. We did not observe differences in the development of common lymphoid progenitor populations. Our findings contrast previous studies but underscore the role of NR2F6 in regulating gene expression levels during mouse bone marrow hematopoiesis and myelopoiesis.
Keywords: GMP; HSC; MDP; bone marrow; hematopoiesis; myeloid compartment; nuclear receptor NR2F6; pre-cDC2.
Copyright © 2025 Woelk, Narasimhan, Pfeifhofer-Obermair, Schraml and Hermann-Kleiter.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

