Mineral Stress Drives Loss of Heterochromatin: An Early Harbinger of Vascular Inflammaging and Calcification
- PMID: 39840455
- PMCID: PMC11825498
- DOI: 10.1161/CIRCRESAHA.124.325374
Mineral Stress Drives Loss of Heterochromatin: An Early Harbinger of Vascular Inflammaging and Calcification
Abstract
Background: Vascular calcification is a detrimental aging pathology markedly accelerated in patients with chronic kidney disease. PLA (prelamin A) is a biomarker of vascular smooth muscle cell aging that accelerates calcification however the mechanisms remain undefined.
Methods: Vascular smooth muscle cells were transduced with PLA using an adenoviral vector and epigenetic modifications were monitored using immunofluorescence and targeted polymerase chain reaction array. Epigenetic findings were verified in vivo using immunohistochemistry in human vessels, in a mouse model of inducible prelamin A expression, and in a rat model of chronic kidney disease-induced calcification. Transcriptomic and chromatin immunoprecipitation followed by sequencing analyses were used to identify gene targets impacted by changes in the epigenetic landscape. Molecular tools and antibody arrays were used to monitor the effects of mineral dysregulation on heterochromatin, inflammation, aging, and calcification.
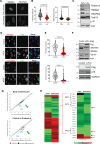
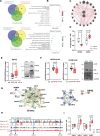
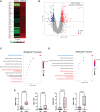
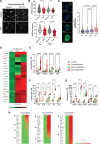
Results: Here, we report that depletion of the repressive heterochromatin marks, H3K9me3 (histone H3, lysine 9, trimethylation) and H3K27me3 (histone H3, lysine 27,trimethylation), is an early hallmark of vascular aging induced by both nuclear lamina dysfunction and dysregulated mineral metabolism, which act to modulate the expression of key epigenetic writers and erasers. Global analysis of H3K9me3 and H3K27me3 marks and pathway analysis revealed deregulation of insulin signaling and autophagy pathways as well as cross-talking DNA damage and NF-κB (nuclear factor κB) inflammatory pathways consistent with early activation of the senescence-associated secretory phenotype. Expression of PLA in vivo induced loss of heterochromatin and promoted inflammation and osteogenic differentiation which preceded aging indices, such as DNA damage and senescence. Vessels from children on dialysis and rats with chronic kidney disease showed prelamin A accumulation and accelerated loss of heterochromatin before the onset of calcification.
Conclusions: Dysregulated mineral metabolism drives changes in the epigenetic landscape and nuclear lamina dysfunction that together promote early induction of inflammaging pathways priming the vasculature for downstream pathological change.
Keywords: aging; heterochromatin; lamins; muscle, smooth, vascular.
Conflict of interest statement
None.
Figures



Comment in
-
Mineral Stress and Vascular Aging: Decoding the Epigenetic Connection to Slow the Clock.Circ Res. 2025 Feb 14;136(4):400-402. doi: 10.1161/CIRCRESAHA.125.326064. Epub 2025 Feb 13. Circ Res. 2025. PMID: 39946443 No abstract available.
References
-
- Sutton NR, Malhotra R, Hilaire CS, Aikawa E, Blumenthal RS, Gackenbach G, Goyal P, Johnson A, Nigwekar SU, Shanahan CM, et al. Molecular mechanisms of vascular health: insights from vascular aging and calcification. Arterioscler Thromb Vasc Biol. 2023;43:15. doi: 10.1161/ATVBAHA.122.317332 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

