Targeting MAPK14 by Lobeline Upregulates Slurp1-Mediated Inhibition of Alternative Activation of TAM and Retards Colorectal Cancer Growth
- PMID: 39840525
- PMCID: PMC11904982
- DOI: 10.1002/advs.202407900
Targeting MAPK14 by Lobeline Upregulates Slurp1-Mediated Inhibition of Alternative Activation of TAM and Retards Colorectal Cancer Growth
Abstract
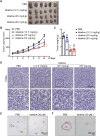
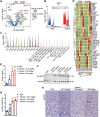
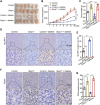
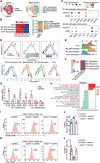
Colorectal cancer (CRC) usually creates an immunosuppressive microenvironment, thereby hindering immunotherapy response. Effective treatment options remain elusive. Using scRNA-seq analysis in a tumor-bearing murine model, it is found that lobeline, an alkaloid from the herbal medicine lobelia, promotes polarization of tumor-associated macrophages (TAMs) toward M1-like TAMs while inhibiting their polarization toward M2-like TAMs. Additionally, lobeline upregulates mRNA expression of secreted Ly-6/UPAR-related protein 1 (Slurp1) in cancer cells. The inhibitory effects of lobeline on tumor load and TAM polarization are almost completely eliminated when Slurp1-deficient MC38 cells are subcutaneously injected into mice, suggesting that lobeline exerts an antitumor effect in a Slurp1-dependent manner. Furthermore, using target-responsive accessibility profiling, MAPK14 is identified as the direct target protein of lobeline. Mechanistically, upon binding to MAPK14 in colon cancer cells, lobeline prevents nuclear translocation of MAPK14, resulting in decreased levels of phosphorylated p53. Consequently, negative transcriptional regulation of SLURP1 by p53 is suppressed, leading to enhanced transcription and secretion of SLURP1. Finally, combination therapy using lobeline and anti-PD1 exhibits stronger antitumor effects. Taken together, these findings suggest that remodeling the immunosuppressive microenvironment using small-molecule lobeline may represent a promising therapeutic strategy for CRC.
Keywords: MAPK14; Slurp1; TAMs polarization; colorectal cancer; lobeline.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


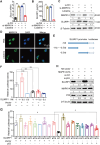
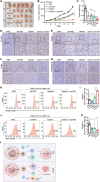
References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J Clin 2021, 71, 209. - PubMed
-
- Ciardiello F., Ciardiello D., Martini G., Napolitano S., Tabernero J., Cervantes A., CA Cancer J Clin 2022, 72, 372. - PubMed
-
- a) Ganesh K., Stadler Z. K., Cercek A., Mendelsohn R. B., Shia J., Segal N. H., L. A. Diaz Jr., Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361; - PMC - PubMed
- b) Ganesh K., Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 93; - PMC - PubMed
- c) Kubli S. P., Berger T., Araujo D. V., Siu L. L., Mak T. W., Nat Rev Drug Discov 2021, 20, 899; - PubMed
- d) Schmitt M., Greten F. R., Nat. Rev. Immunol. 2021, 21, 653; - PubMed
- e) Tie Y., Tang F., Wei Y. Q., Wei X. W., J. Hematol. Oncol. 2022, 15, 61. - PMC - PubMed
-
- a) Strauss L., Mahmoud M. A. A., Weaver J. D., Tijaro‐Ovalle N. M., Christofides A., Wang Q., Pal R., Yuan M., Asara J., Patsoukis N., Boussiotis V. A., Sci Immunol 2020, 5, eaay1863; - PMC - PubMed
- b) Dixon K. O., Tabaka M., Schramm M. A., Xiao S., Tang R., Dionne D., Anderson A. C., Rozenblatt‐Rosen O., Regev A., Kuchroo V. K., NatureNature 2021, 595, 101; - PMC - PubMed
- c) Seo W. I., Lee C. H., Jung S. J., Lee D. S., Park H. Y., Jeong D. H., Kim W., Chung J. I., Choi I., Cancer Immunol. Immunother. 2021, 70, 3113; - PMC - PubMed
- d) Veglia F., Sanseviero E., Gabrilovich D. I., Nat. Rev. Immunol. 2021, 21, 485. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC3500202/National Key Research and Development Plan
- 82304799/National Natural Science Foundation of China
- U21A20345/National Natural Science Foundation of China
- 020814380179/Fundamental Research Funds for the Central Universities
- 020814380174/Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
