Translation of the downstream ORF from bicistronic mRNAs by human cells: Impact of codon usage and splicing in the upstream ORF
- PMID: 39840808
- PMCID: PMC11751868
- DOI: 10.1002/pro.70036
Translation of the downstream ORF from bicistronic mRNAs by human cells: Impact of codon usage and splicing in the upstream ORF
Abstract
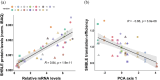
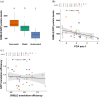
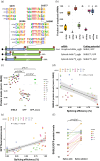
Biochemistry textbooks describe eukaryotic mRNAs as monocistronic. However, increasing evidence reveals the widespread presence and translation of upstream open reading frames preceding the "main" ORF. DNA and RNA viruses infecting eukaryotes often produce polycistronic mRNAs and viruses have evolved multiple ways of manipulating the host's translation machinery. Here, we introduce an experimental model to study gene expression regulation from virus-like bicistronic mRNAs in human cells. The model consists of a short upstream ORF and a reporter downstream ORF encoding a fluorescent protein. We have engineered synonymous variants of the upstream ORF to explore large parameter space, including codon usage preferences, mRNA folding features, and splicing propensity. We show that human translation machinery can translate the downstream ORF from bicistronic mRNAs, albeit reporter protein levels are thousand times lower than those from the upstream ORF. Furthermore, synonymous recoding of the upstream ORF exclusively during elongation significantly influences its own translation efficiency, reveals cryptic splice signals, and modulates the probability of downstream ORF translation. Our results are consistent with a leaky scanning mechanism facilitating downstream ORF translation from bicistronic mRNAs in human cells, offering new insights into the role of upstream ORFs in translation regulation.
Keywords: GFP; RNA splicing; bicistronic mRNA; codon usage; downstream ORF; elongation; eukaryote; fluorescence; main ORF; polycistronic mRNA; transcription; translation; upstream ORF.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Alexaki A, Kames J, Holcomb DD, Athey J, Santana‐Quintero LV, Lam PVN, et al. Codon and codon‐pair usage tables (CoCoPUTs): facilitating genetic variation analyses and recombinant gene design. J Mol Biol. 2019;431:2434–2441. - PubMed
-
- Arike L, Valgepea K, Peil L, Nahku R, Adamberg K, Vilu R. Comparison and applications of label‐free absolute proteome quantification methods on Escherichia coli . J Proteomics. 2012;75:5437–5448. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

