Combining computational modeling and experimental library screening to affinity-mature VEEV-neutralizing antibody F5
- PMID: 39840828
- PMCID: PMC11752144
- DOI: 10.1002/pro.70043
Combining computational modeling and experimental library screening to affinity-mature VEEV-neutralizing antibody F5
Abstract
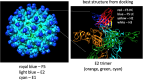
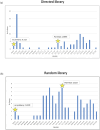
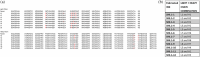
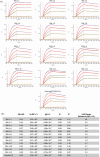
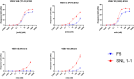
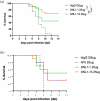
Engineered monoclonal antibodies have proven to be highly effective therapeutics in recent viral outbreaks. However, despite technical advancements, an ability to rapidly adapt or increase antibody affinity and by extension, therapeutic efficacy, has yet to be fully realized. We endeavored to stand-up such a pipeline using molecular modeling combined with experimental library screening to increase the affinity of F5, a monoclonal antibody with potent neutralizing activity against Venezuelan Equine Encephalitis Virus (VEEV), to recombinant VEEV (IAB) E1E2 antigen. We modeled the F5/E1E2 binding interface and generated predictions for mutations to improve binding using a Rosetta-based approach and dTERMen, an informatics approach. The modeling was complicated by the fact that a high-resolution structure of F5 is not available and the H3 loop of F5 exceeds the length for which current modeling approaches can determine a unique structure. A subset of the predicted mutations from both methods were incorporated into a phage display library of scFvs. This library and a library generated by error-prone PCR were screened for binding affinity to the recombinant antigen. Results from the screens identified favorable mutations which were incorporated into 12 human-IgG1 variants. The best variant, containing eight mutations, improved KD from 0.63 nM (parental) to 0.01 nM. While this did not improve neutralization or therapeutic potency of F5 against IAB, it did increase cross-reactivity to other closely related VEEV epizootic and enzootic strains, demonstrating the potential of this method to rapidly adapt existing therapeutics to emerging viral strains.
Keywords: affinity‐maturation; anti‐viral; computational modeling; library screening; therapeutic antibodies.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

