Exploring animal food microbiomes and resistomes via 16S rRNA gene amplicon sequencing and shotgun metagenomics
- PMID: 39840975
- PMCID: PMC11837513
- DOI: 10.1128/aem.02230-24
Exploring animal food microbiomes and resistomes via 16S rRNA gene amplicon sequencing and shotgun metagenomics
Abstract
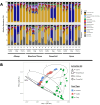
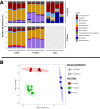
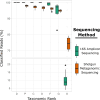
As a diverse and complex food matrix, the animal food microbiota and repertoire of antimicrobial resistance (AMR) genes remain to be better understood. In this study, 16S rRNA gene amplicon sequencing and shotgun metagenomics were applied to three types of animal food samples (cattle feed, dry dog food, and poultry feed). ZymoBIOMICS mock microbial community was used for workflow optimization including DNA extraction kits and bead-beating conditions. The four DNA extraction kits (AllPrep PowerViral DNA/RNA Kit, DNeasy Blood & Tissue Kit, DNeasy PowerSoil Kit, and ZymoBIOMICS DNA Miniprep Kit) were compared in animal food as well as the use of peptide nucleic acid blockers for 16S rRNA gene amplicon sequencing. Distinct microbial community profiles were generated, which varied by animal food type and DNA extraction kit. Employing peptide nucleic acid blockers prior to 16S rRNA gene amplicon sequencing was comparable with post-sequencing in silico filtering at removing chloroplast and mitochondrial sequences. There was a good agreement between 16S rRNA gene amplicon sequencing and shotgun metagenomics on community profiles in animal feed data sets; however, they differed in taxonomic resolution, with the latter superior at resolving at the species level. Although the overall prevalence of AMR genes was low, resistome analysis of animal feed data sets by shotgun metagenomics revealed 10 AMR gene/protein families, including beta-lactamases, erythromycin/lincomycin/pristinamycin/tylosin, fosfomycin, phenicol, and quinolone. Future expansion of microbiome and resistome profiling in animal food will help better understand the bacterial and AMR gene diversity in these commodities and help guide pathogen control and AMR prevention efforts.IMPORTANCEWith the growing interest and application of metagenomics in understanding the structure/composition and function of diverse microbial communities along the One Health continuum, this study represents one of the first attempts to employ these advanced sequencing technologies to characterize the microbiota and AMR genes in animal food. We unraveled the effects of DNA extraction kits on sample analysis by 16S rRNA gene amplicon sequencing and showed similar efficacies of two strategies at removing chloroplast and mitochondrial reads. The in-depth analysis using shotgun metagenomics shed light on the community compositions and the presence of an array of AMR genes in animal food. This exploration of microbiomes and resistomes in representative animal food samples by both sequencing approaches laid important groundwork for future metagenomic investigations to gain a better understanding of the baseline/core microbiomes and associated AMR functions in these diverse commodities and help guide pathogen control and AMR prevention efforts.
Keywords: 16S rRNA gene amplicon sequencing; DNA extraction; animal food; microbiome; mock community; peptide nucleic acid; resistome; shotgun metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evaluation of DNA extraction kits for long-read shotgun metagenomics using Oxford Nanopore sequencing for rapid taxonomic and antimicrobial resistance detection.Sci Rep. 2024 Nov 27;14(1):29531. doi: 10.1038/s41598-024-80660-3. Sci Rep. 2024. PMID: 39604411 Free PMC article.
-
Impact of DNA extraction, PCR amplification, sequencing, and bioinformatic analysis on food-associated mock communities using PacBio long-read amplicon sequencing.BMC Microbiol. 2024 Dec 6;24(1):521. doi: 10.1186/s12866-024-03677-8. BMC Microbiol. 2024. PMID: 39643893 Free PMC article.
-
Investigation of tylosin in feed of feedlot cattle and effects on liver abscess prevalence, and fecal and soil microbiomes and resistomes1.J Anim Sci. 2019 Nov 4;97(11):4567-4578. doi: 10.1093/jas/skz306. J Anim Sci. 2019. PMID: 31563955 Free PMC article.
-
Do we need a standardized 16S rRNA gene amplicon sequencing analysis protocol for poultry microbiota research?Poult Sci. 2025 Jul;104(7):105242. doi: 10.1016/j.psj.2025.105242. Epub 2025 May 1. Poult Sci. 2025. PMID: 40334389 Free PMC article. Review.
-
Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments.J Microbiol Methods. 2021 May;184:106174. doi: 10.1016/j.mimet.2021.106174. Epub 2021 Mar 24. J Microbiol Methods. 2021. PMID: 33774111 Free PMC article. Review.
References
-
- Alltech . 2024. 2024 alltech agri-food outlook. Available from: https://www.alltech.com/press-release/2024-alltech-agri-food-outlook-sha.... Retrieved 26 Nov 2024.
-
- Decision Innovation Solutions . 2023. Economic contribution of the animal feed and pet food manufacturing industries. Available from: https://www.afia.org/feedfacts/feed-industry-stats/economic-impact. Retrieved 26 Nov 2024.
-
- FDA . 2023. 21 CFR subchapter E: animal drugs, feeds, and related products (parts 500-599). Available from: https://www.govinfo.gov/content/pkg/CFR-2023-title21-vol6/pdf/CFR-2023-t.... Retrieved 26 Nov 2024.
-
- EC . 2024. Animal feed. Available from: https://ec.europa.eu/food/safety/animal-feed_en. Retrieved 26 Nov 2024.
-
- FDA . 2024. Animal food & feeds. Available from: https://www.fda.gov/animal-veterinary/products/animal-food-feeds. Retrieved 26 Nov 2024.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

