Novel clinical trial designs emerging from the molecular reclassification of cancer
- PMID: 39841128
- PMCID: PMC12061631
- DOI: 10.3322/caac.21880
Novel clinical trial designs emerging from the molecular reclassification of cancer
Abstract
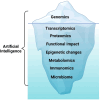
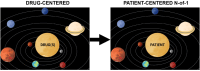
Next-generation sequencing has revealed the disruptive reality that advanced/metastatic cancers have complex and individually distinct genomic landscapes, necessitating a rethinking of treatment strategies and clinical trial designs. Indeed, the molecular reclassification of cancer suggests that it is the molecular underpinnings of the disease, rather than the tissue of origin, that mostly drives outcomes. Consequently, oncology clinical trials have evolved from standard phase 1, 2, and 3 tissue-specific studies; to tissue-specific, biomarker-driven trials; to tissue-agnostic trials untethered from histology (all drug-centered designs); and, ultimately, to patient-centered, N-of-1 precision medicine studies in which each patient receives a personalized, biomarker-matched therapy/combination of drugs. Innovative technologies beyond genomics, including those that address transcriptomics, immunomics, proteomics, functional impact, epigenetic changes, and metabolomics, are enabling further refinement and customization of therapy. Decentralized studies have the potential to improve access to trials and precision medicine approaches for underserved minorities. Evaluation of real-world data, assessment of patient-reported outcomes, use of registry protocols, interrogation of exceptional responders, and exploitation of synthetic arms have all contributed to personalized therapeutic approaches. With greater than 1 × 1012 potential patterns of genomic alterations and greater than 4.5 million possible three-drug combinations, the deployment of artificial intelligence/machine learning may be necessary for the optimization of individual therapy and, in the near future, also may permit the discovery of new treatments in real time.
Keywords: N‐of‐1; cancer; personalized medicine; precision medicine; tissue agnostic.
© 2025 The Author(s). CA: A Cancer Journal for Clinicians published by Wiley Periodicals LLC on behalf of American Cancer Society.
Conflict of interest statement
Shumei Kato reports research funding from ACT Genomics, Sysmex, Konica Minolta, OmniSeq, Personalis, and Function Oncology; personal/consulting fees from Medpace, Foundation Medicine, NeoGenomics and CureMatch; speaker's fees from Chugai, Roche/Genentech, and Bayer; and service on an advisory board for Pfizer outside the submitted work. Jason K. Sicklick reports consulting fees from Deciphera, Kura, and CureMatch; speaker's fees from Deciphera, La‐Hoffman Roche, Foundation Medicine, Merck, QED, Daiichi Sankyo, and SpringWorks; and owns stock in Personalis and CureMatch outside the submitted work. Razelle Kurzrock reports research funding from Boehringer Ingelheim, the National Cancer Institute, Debiopharm, Foundation Medicine, Genentech, Grifols, Guardant, Incyte, Konica Minolta, Medimmune, Merck Serono, Omniseq, Pfizer, Sequenom, Takeda, and TopAlliance; consulting, speaker's, and/or advisory board fees from Actuate Therapeutics, AstraZeneca, Bicara Therapeutics Inc., Biological Dynamics, Caris, Datar Cancer Genetics, Daiichi, EISAI, EOM Pharmaceuticals, Iylon, LabCorp, Merck, NeoGenomics, Neomed, Pfizer, Precirix, Prosperdtx, Regeneron, Roche, TD2/Volastra, Turning Point Therapeutics, and X‐Biotech; equity interest in CureMatch Inc. and IDbyDNA; service on the boards of CureMatch and CureMetrix; and is a co‐founder of CureMatch outside the submitted work. The remaining authors disclosed no conflicts of interest.
Figures
Similar articles
-
Clinical trial design in the era of precision medicine.Genome Med. 2022 Aug 31;14(1):101. doi: 10.1186/s13073-022-01102-1. Genome Med. 2022. PMID: 36045401 Free PMC article. Review.
-
Worldwide Innovative Network (WIN) Consortium in Personalized Cancer Medicine: Bringing next-generation precision oncology to patients.Oncotarget. 2025 Mar 12;16:140-162. doi: 10.18632/oncotarget.28703. Oncotarget. 2025. PMID: 40073368 Free PMC article.
-
Personalized Medicine: Genomics Trials in Oncology.Trans Am Clin Climatol Assoc. 2015;126:133-43. Trans Am Clin Climatol Assoc. 2015. PMID: 26330667 Free PMC article. Review.
-
From Tissue-Agnostic to N-of-One Therapies: (R)Evolution of the Precision Paradigm.Trends Cancer. 2021 Jan;7(1):15-28. doi: 10.1016/j.trecan.2020.08.009. Epub 2020 Sep 29. Trends Cancer. 2021. PMID: 33008795 Review.
-
Review of precision cancer medicine: Evolution of the treatment paradigm.Cancer Treat Rev. 2020 Jun;86:102019. doi: 10.1016/j.ctrv.2020.102019. Epub 2020 Mar 31. Cancer Treat Rev. 2020. PMID: 32251926 Free PMC article. Review.
Cited by
-
Nectin-4 expression patterns and therapeutics in oncology.Cancer Lett. 2025 Jul 10;622:217681. doi: 10.1016/j.canlet.2025.217681. Epub 2025 Apr 8. Cancer Lett. 2025. PMID: 40209851 Review.
-
Bridging epigenomics and tumor immunometabolism: molecular mechanisms and therapeutic implications.Mol Cancer. 2025 Mar 8;24(1):71. doi: 10.1186/s12943-025-02269-y. Mol Cancer. 2025. PMID: 40057791 Free PMC article. Review.
-
Tumor-Agnostic Therapies in Practice: Challenges, Innovations, and Future Perspectives.Cancers (Basel). 2025 Feb 26;17(5):801. doi: 10.3390/cancers17050801. Cancers (Basel). 2025. PMID: 40075649 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

