Intercellular mRNA transfer alters the human pluripotent stem cell state
- PMID: 39841146
- PMCID: PMC11789055
- DOI: 10.1073/pnas.2413351122
Intercellular mRNA transfer alters the human pluripotent stem cell state
Abstract
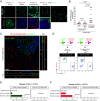
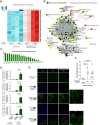
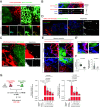
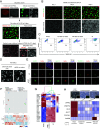
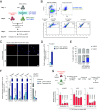
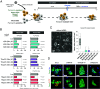
Intercellular transmission of messenger RNA (mRNA) is being explored in mammalian species using immortal cell lines. Here, we uncover an intercellular mRNA transfer phenomenon that allows for the adaptation and reprogramming of human primed pluripotent stem cells (hPSCs). This process is induced by the direct cell contact-mediated coculture with mouse embryonic stem cells under the condition impermissible for primed hPSC culture. Mouse-derived mRNA contents are transmitted into adapted hPSCs only in the coculture. Transfer-specific mRNA analysis shows the enrichment for divergent biological pathways involving transcription/translational machinery and stress-coping mechanisms, wherein such transfer is diminished when direct cell contacts are lost. After 5 d of coculture with mouse embryonic stem cells, surface marker analysis and global gene profiling confirmed that mRNA transfer-prone hPSC efficiently gains a naïve-like state. Furthermore, transfer-specific knockdown experiments targeting mouse-specific transcription factor-coding mRNAs in hPSC show that mouse-derived Tfcp2l1, Tfap2c, and Klf4 are indispensable for human naïve-like conversion. Thus, interspecies mRNA transfer triggers cellular reprogramming in mammalian cells. Our results support that episodic mRNA transfer can occur in cell cooperative and competitive processes, which provides a fresh perspective on understanding the roles of mRNA mobility for intra- and interspecies cellular communications.
Keywords: cell–cell communication; mRNA transfer; pluripotency; reprogramming.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
Update of
-
Inter-cellular mRNA Transfer Alters Human Pluripotent Stem Cell State.bioRxiv [Preprint]. 2024 Jun 27:2024.06.27.600209. doi: 10.1101/2024.06.27.600209. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Jan 28;122(4):e2413351122. doi: 10.1073/pnas.2413351122. PMID: 38979277 Free PMC article. Updated. Preprint.
Similar articles
-
Inter-cellular mRNA Transfer Alters Human Pluripotent Stem Cell State.bioRxiv [Preprint]. 2024 Jun 27:2024.06.27.600209. doi: 10.1101/2024.06.27.600209. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Jan 28;122(4):e2413351122. doi: 10.1073/pnas.2413351122. PMID: 38979277 Free PMC article. Updated. Preprint.
-
Derivation of trophoblast stem cells from naïve human pluripotent stem cells.Elife. 2020 Feb 12;9:e52504. doi: 10.7554/eLife.52504. Elife. 2020. PMID: 32048992 Free PMC article.
-
STAT3-dependent long non-coding RNA Lncenc1 contributes to mouse ES cells pluripotency via stabilizing Klf4 mRNA.Brief Funct Genomics. 2024 Sep 27;23(5):651-662. doi: 10.1093/bfgp/elad045. Brief Funct Genomics. 2024. PMID: 37801430 Free PMC article.
-
Moving toward totipotency: the molecular and cellular features of totipotent and naive pluripotent stem cells.Hum Reprod Update. 2025 Jul 1;31(4):361-391. doi: 10.1093/humupd/dmaf006. Hum Reprod Update. 2025. PMID: 40299455 Review.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
Cited by
-
Mast cells interact directly with colorectal cancer cells to promote epithelial-to-mesenchymal transition.bioRxiv [Preprint]. 2025 Mar 19:2025.03.19.644113. doi: 10.1101/2025.03.19.644113. bioRxiv. 2025. PMID: 40166179 Free PMC article. Preprint.
-
Energy Metabolism and Stemness and the Role of Lauric Acid in Reversing 5-Fluorouracil Resistance in Colorectal Cancer Cells.Int J Mol Sci. 2025 Jan 14;26(2):664. doi: 10.3390/ijms26020664. Int J Mol Sci. 2025. PMID: 39859378 Free PMC article.
References
-
- Valadi H., et al. , Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007). - PubMed
-
- Jiang H., Li Z., Li X., Xia J., Intercellular transfer of messenger RNAs in multiorgan tumorigenesis by tumor cell-derived exosomes. Mol. Med. Rep. 11, 4657–4663 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

