Chemical tools to define and manipulate interferon-inducible Ubl protease USP18
- PMID: 39843430
- PMCID: PMC11754618
- DOI: 10.1038/s41467-025-56336-5
Chemical tools to define and manipulate interferon-inducible Ubl protease USP18
Abstract
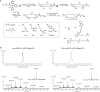
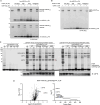
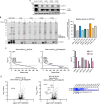
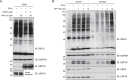
Ubiquitin-specific protease 18 (USP18) is a multifunctional cysteine protease primarily responsible for deconjugating the interferon-inducible ubiquitin-like modifier ISG15 from protein substrates. Here, we report the design and synthesis of activity-based probes (ABPs) that incorporate unnatural amino acids into the C-terminal tail of ISG15, enabling the selective detection of USP18 activity over other ISG15 cross-reactive deubiquitinases (DUBs) such as USP5 and USP14. Combined with a ubiquitin-based DUB ABP, the USP18 ABP is employed in a chemoproteomics screening platform to identify and assess inhibitors of DUBs including USP18. We further demonstrate that USP18 ABPs can be utilized to profile differential activities of USP18 in lung cancer cell lines, providing a strategy that will help define the activity-related landscape of USP18 in different disease states and unravel important (de)ISGylation-dependent biological processes.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Update of
-
Chemical tools to define and manipulate interferon-inducible Ubl protease USP18.bioRxiv [Preprint]. 2024 Apr 8:2024.04.08.588544. doi: 10.1101/2024.04.08.588544. bioRxiv. 2024. Update in: Nat Commun. 2025 Jan 22;16(1):957. doi: 10.1038/s41467-025-56336-5. PMID: 38645224 Free PMC article. Updated. Preprint.
Similar articles
-
Chemical tools to define and manipulate interferon-inducible Ubl protease USP18.bioRxiv [Preprint]. 2024 Apr 8:2024.04.08.588544. doi: 10.1101/2024.04.08.588544. bioRxiv. 2024. Update in: Nat Commun. 2025 Jan 22;16(1):957. doi: 10.1038/s41467-025-56336-5. PMID: 38645224 Free PMC article. Updated. Preprint.
-
Emerging Roles of USP18: From Biology to Pathophysiology.Int J Mol Sci. 2020 Sep 17;21(18):6825. doi: 10.3390/ijms21186825. Int J Mol Sci. 2020. PMID: 32957626 Free PMC article. Review.
-
Structural basis of the specificity of USP18 toward ISG15.Nat Struct Mol Biol. 2017 Mar;24(3):270-278. doi: 10.1038/nsmb.3371. Epub 2017 Feb 6. Nat Struct Mol Biol. 2017. PMID: 28165509 Free PMC article.
-
Mechanisms of USP18 specificity toward ISG15 revealed by paralog sequence analysis comparison.J Biol Chem. 2025 Jul;301(7):110288. doi: 10.1016/j.jbc.2025.110288. Epub 2025 May 26. J Biol Chem. 2025. PMID: 40436319 Free PMC article.
-
USP18-mediated protein deISGylation and its role in tuberculosis and other infectious diseases.Yi Chuan. 2023 Nov 20;45(11):998-1006. doi: 10.16288/j.yczz.23-185. Yi Chuan. 2023. PMID: 38764265 Review.
References
-
- Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol.10, 550–563 (2009). - PubMed
-
- Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D. E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem.277, 9976–9981 (2002). - PubMed
-
- Narasimhan, J., Potter, J. L. & Haas, A. L. Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem.271, 324–330 (1996). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

