Telomerase RNA structural heterogeneity in living human cells detected by DMS-MaPseq
- PMID: 39843442
- PMCID: PMC11754830
- DOI: 10.1038/s41467-025-56149-6
Telomerase RNA structural heterogeneity in living human cells detected by DMS-MaPseq
Abstract
Biogenesis of human telomerase requires its RNA subunit (hTR) to fold into a multi-domain architecture that includes the template-pseudoknot (t/PK) and the three-way junction (CR4/5). These hTR domains bind the telomerase reverse transcriptase (hTERT) protein and are essential for telomerase activity. Here, we probe hTR structure in living cells using dimethyl sulfate mutational profiling with sequencing (DMS-MaPseq) and ensemble deconvolution analysis. Approximately 15% of the steady state population of hTR has a CR4/5 conformation lacking features required for hTERT binding. The proportion of hTR CR4/5 folded into the primary functional conformation is independent of hTERT expression levels. Mutations that stabilize the alternative CR4/5 conformation are detrimental to telomerase assembly and activity. Moreover, the alternative CR4/5 conformation is not found in purified telomerase RNP complexes, supporting the hypothesis that only the primary CR4/5 conformer is active. We propose that this misfolded portion of the cellular hTR pool is either slowly refolded or degraded, suggesting that kinetic RNA folding traps studied in vitro may also hinder ribonucleoprotein assembly in vivo.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.R.C. is a scientific advisor for Eikon Therapeutics, Storm Therapeutics, lincSwitch Therapeutics, and Somalogic, Inc. The remaining authors declare no competing interests.
Figures



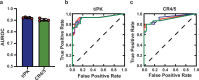

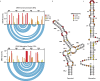
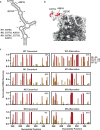



Update of
-
Telomerase RNA structural heterogeneity in living human cells detected by DMS-MaPseq.bioRxiv [Preprint]. 2024 Aug 29:2023.10.04.560962. doi: 10.1101/2023.10.04.560962. bioRxiv. 2024. Update in: Nat Commun. 2025 Jan 22;16(1):925. doi: 10.1038/s41467-025-56149-6. PMID: 37873413 Free PMC article. Updated. Preprint.
References
-
- de Lange, T. Shelterin-mediated telomere protection. Annu Rev. Genet52, 223–247 (2018). - PubMed
-
- Greider, C. W. & Blackburn, E. H. Tracking telomerase. Cell116, S83–S86 (2004). 81 p following S86. - PubMed
-
- Harley, C. B., Futcher, A. B. & Greider, C. W. Telomeres shorten during ageing of human fibroblasts. Nature345, 458–460 (1990). - PubMed
-
- Olovnikov, A. M. Telomeres, telomerase, and aging: origin of the theory. Exp. Gerontol.31, 443–448 (1996). - PubMed
MeSH terms
Substances
Grants and funding
- R01 GM095850/GM/NIGMS NIH HHS/United States
- R01GM095850/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 GM133391/GM/NIGMS NIH HHS/United States
- R35 GM153235/GM/NIGMS NIH HHS/United States
- R35GM153235/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)

