A legume-enriched diet improves metabolic health in prediabetes mediated through gut microbiome: a randomized controlled trial
- PMID: 39843443
- PMCID: PMC11754483
- DOI: 10.1038/s41467-025-56084-6
A legume-enriched diet improves metabolic health in prediabetes mediated through gut microbiome: a randomized controlled trial
Abstract
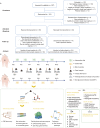
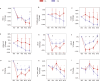
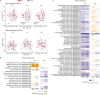
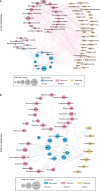
Healthy dietary patterns rich in legumes can improve metabolic health, although their additional benefits in conjunction with calorie restriction have not been well-established. We investigated effects of a calorie-restricted, legume-enriched, multicomponent intervention diet compared with a calorie-restricted control diet in 127 Chinese prediabetes participants, living in Singapore. The study was a 16-week, single-blind, parallel-design, randomized controlled trial (n = 63 intervention group (IG), n = 64 control group (CG); mean ± SD age 62.2 ± 6.3 years, BMI 23.8 ± 2.6 kg/m2). Primary outcomes were markers of glycemia and all measurements were taken at 2 or 4-weekly intervals. At the end of 16 weeks, both groups had significantly lower BMI (q(Time) = 1.92 ×10-42, β = -0.02) compared with baseline, with minimal difference between groups. The IG had significantly greater reductions in LDL cholesterol (q(Treatment×Time) = 0.01, β = -0.16), total cholesterol (q(Treatment×Time) = 0.02, β = -0.3) and HbA1c (q(Treatment×Time) = 0.04, β = -0.004) compared with CG, alongside increases in fiber degrading species in IG, mediated through metabolites such as bile acids and amino acids. A legume-enriched, multicomponent intervention diet can improve metabolic health in a prediabetes population, in addition to benefits obtained from calorie restriction alone, partially mediated through changes in gut microbial composition and function. Trial registration: Clinical Trials NCT04745702.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: X.W., K.J.L., V.S.B.V., K.K., X.L., L.H.W., C.P.L.Y. and H.F. are employees of Wilmar International Limited. The remaining authors declare no competing interests. Inclusion & ethics: This study aligns by the guidelines of Nature Portfolio journals, with contributions of each author as disclosed in Author contributions.
Figures



References
-
- Saeedi, P. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabet. Res. Clin. Pract.157, 107843 (2019). - PubMed
-
- Esposito, K. et al. Which diet for prevention of type 2 diabetes? A meta-analysis of prospective studies. Endocrine47, 107–116 (2014). - PubMed
-
- Kris-Etherton, P. M., Etherton, T. D., Carlson, J. & Gardner, C. Recent discoveries in inclusive food-based approaches and dietary patterns for reduction in risk for cardiovascular disease. Curr. Opin. Lipido.13, 397–407 (2002). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

