Antibiotic resistance mediated by gene amplifications
- PMID: 39843582
- PMCID: PMC11721125
- DOI: 10.1038/s44259-024-00052-5
Antibiotic resistance mediated by gene amplifications
Abstract
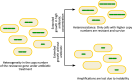
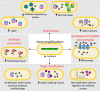
Apart from horizontal gene transfer and sequence-altering mutational events, antibiotic resistance can emerge due to the formation of tandem repeats of genomic regions. This phenomenon, also known as gene amplification, has been implicated in antibiotic resistance in both laboratory and clinical scenarios, where the evolution of resistance via amplifications can affect treatment efficacy. Antibiotic resistance mediated by gene amplifications is unstable and consequently can be difficult to detect, due to amplification loss in the absence of the selective pressure of the antibiotic. Further, due to variable copy numbers in a population, amplifications result in heteroresistance, where only a subpopulation is resistant to an antibiotic. While gene amplifications typically lead to resistance by increasing the expression of resistance determinants due to the higher copy number, the underlying mechanisms of resistance are diverse. In this review article, we describe the various pathways by which gene amplifications cause antibiotic resistance, from efflux and modification of the antibiotic, to target modification and bypass. We also discuss how gene amplifications can engender resistance by alternate mutational outcomes such as altered regulation and protein structure, in addition to just an increase in copy number and expression. Understanding how amplifications contribute to bacterial survival following antibiotic exposure is critical to counter their role in the rise of antimicrobial resistance.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- CDC. Antibiotic Resistance Threats in the United States (AR Threats Report). (2019).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

