Identification of promising SARS-CoV-2 main protease inhibitor through molecular docking, dynamics simulation, and ADMET analysis
- PMID: 39843610
- PMCID: PMC11754916
- DOI: 10.1038/s41598-025-86016-9
Identification of promising SARS-CoV-2 main protease inhibitor through molecular docking, dynamics simulation, and ADMET analysis
Abstract
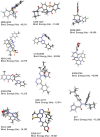
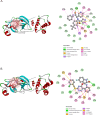
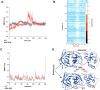
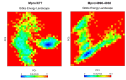
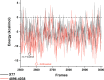
The COVID-19 pandemic caused by SARS-CoV-2 continues to pose a major challenge to global health. Targeting the main protease of the virus (Mpro), which is essential for viral replication and transcription, offers a promising approach for therapeutic intervention. In this study, advanced computational techniques such as molecular docking and molecular dynamics simulations were used to screen a series of antiviral compounds for their potential inhibitory effect on the SARS-CoV-2 Mpro. A comprehensive analysis of compounds from the ChemDiv and PubChem databases was performed. The physicochemical properties, pharmacokinetics, and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) profiles were evaluated to determine drug similarity and safety. Compound 4896 - 4038 proved to be the most promising candidate. It exhibited a favorable balance between molecular weight (491.06) and lipophilicity (logP 3.957), high intestinal absorption (92.119%), and broad tissue distribution (VDss of 0.529), indicating good oral bioavailability and therapeutic potential. Molecular docking studies showed that 4896 - 4038 has a strong binding affinity to the active site of Mpro and forms key interactions, such as hydrogen bonds, carbon-hydrogen bonds, pi-sulfur, and multiple van der Waals and pi-pi stacked bonds. The binding energy was comparable to that of the reference drug X77, indicating potential efficacy. Molecular dynamics simulations over 300 ns confirmed the stability of the Mpro/4896 - 4038 complex of protein-ligand. Free energy landscape mapping and MM/PBSA calculations further substantiated the favorable binding and stability of the complex. Importantly, 4896 - 4038 exhibited a comparatively favorable safety profile. In summary, compound 4896 - 4038 shows significant potential as a potent SARS-CoV-2 Mpro inhibitor, combining potent inhibitory activity with favorable pharmacokinetic and safety profiles. These results support the further development of 4896 - 4038 as a promising therapeutic agent in the fight against COVID-19 that warrants experimental validation and clinical investigation.
Keywords: 3CLpro; COVID-19; Molecular Docking; Molecular dynamics, ADMET; Mpro; SARS-CoV-2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Ciotti, M. et al. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci.57, 365–388. 10.1080/10408363.2020.1783198 (2020). - PubMed
-
- BS, M. & Nambiar, V. COVID-19: an insight into SARS-CoV-2 Pandemic originated at Wuhan City in Hubei Province of China., 10.23937/2474-3658/1510146
-
- Wambua, D. Human Rights Dimension of a Pandemic: Evaluation of the Effect of Covid 19 on the Right to Education in Kenya. Thesis, University of Nairobi, (2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

