Unraveling the distinctive gut microbiome of khulans (Equus hemionus hemionus) in comparison to their drinking water and closely related equids
- PMID: 39843625
- PMCID: PMC11754619
- DOI: 10.1038/s41598-025-87216-z
Unraveling the distinctive gut microbiome of khulans (Equus hemionus hemionus) in comparison to their drinking water and closely related equids
Abstract
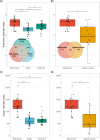
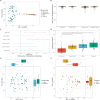
The microbial composition of host-associated microbiomes is influenced by co-evolutionary interactions, host genetics, domestication, and the environment. This study investigates the contribution of environmental microbiota from freshwater bodies to the gastrointestinal microbiomes of wild khulans (Equus hemionus hemionus, n = 21) and compares them with those of captive khulans (n = 12) and other equids-Przewalski's horse (n = 82) and domestic horse (n = 26). Using PacBio technology and the LotuS pipeline for 16S rRNA gene sequencing, we analyze microbial diversity and conduct differential abundance, alpha, and beta diversity analyses. Results indicate limited microbial sharing between wild khulans and their waterhole environments, suggesting minimal environmental influence on their gut microbiomes and low levels of water contamination by khulans. Wild khulans exhibit greater microbial diversity and richness compared to captive ones, likely due to adaptations to the harsh nutritional conditions of the Gobi desert. Conversely, captive khulans show reduced microbial diversity, potentially affected by dietary changes during captivity. These findings highlight the significant impact of environment and lifestyle on the gut microbiomes of equids.
Keywords: Equus hemionus hemionus; 16S rRNA full length gene sequencing; Freshwater microbiota; Gobi Desert; Microbiome; PacBio.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Numberger, D. et al. Urbanization promotes specific bacteria in freshwater microbiomes including potential pathogens. Sci. Total Environ.845, 157321 (2022). - PubMed
-
- Dayaram, A. et al. Seasonal host and ecological drivers may promote restricted water as a viral vector. Sci. Total Environ.773, 145446 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

