FABP4 as a therapeutic host target controlling SARS-CoV-2 infection
- PMID: 39843629
- PMCID: PMC11904229
- DOI: 10.1038/s44321-024-00188-x
FABP4 as a therapeutic host target controlling SARS-CoV-2 infection
Abstract
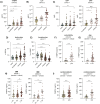
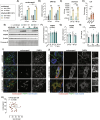
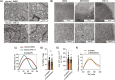
Host metabolic fitness is a critical determinant of infectious disease outcomes. Obesity, aging, and other related metabolic disorders are recognized as high-risk disease modifiers for respiratory infections, including coronavirus infections, though the underlying mechanisms remain unknown. Our study highlights fatty acid-binding protein 4 (FABP4), a key regulator of metabolic dysfunction and inflammation, as a modulator of SARS-CoV-2 pathogenesis, correlating strongly with disease severity in COVID-19 patients. We demonstrate that loss of FABP4 function, by genetic or pharmacological means, reduces SARS-CoV-2 replication and disrupts the formation of viral replication organelles in adipocytes and airway epithelial cells. Importantly, FABP4 inhibitor treatment of infected hamsters diminished lung viral titers, alleviated lung damage and reduced collagen deposition. These findings highlight the therapeutic potential of targeting host metabolism in limiting coronavirus replication and mitigating the pathogenesis of infection.
Keywords: COVID-19; FABP4; Lipid Droplets; Replication Organelles; SARS-CoV-2.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. Emre Koyuncu is co-founder, director, and officer of Crescenta Biosciences and holds equity at the company. Ali Al-Hakeem and Fatih Semerci are employees of Crescenta Biosciences. Hahn Kim is co-founder, director, and consultant of Crescenta Biosciences and holds equity at the company. Hahn Kim is an employee of Princeton University; All work of Hahn Kim included herein was performed as a consultant for Crescenta, independent of Princeton University. Hahn Kim, Emre Koyuncu and Gökhan S. Hotamisligil are inventors on patent application that includes CRE-14. Gökhan S. Hotamisligil is a scientific advisor, receives compensation and holds equity at Crescenta Biosciences. He is also a member of the journal’s advisory editorial board. This has no bearing on the editorial consideration of this article for publication. Other authors declare no competing interests.
Figures



References
-
- Beucher G, Blondot ML, Celle A, Pied N, Recordon-Pinson P, Esteves P, Faure M, Métifiot M, Lacomme S, Dacheux D et al (2022) Bronchial epithelia from adults and children: SARS-CoV-2 spread via syncytia formation and type III interferon infectivity restriction. Proc Natl Acad Sci USA 119:e2202370119 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

