YgiV promoter mutations cause resistance to cystobactamids and reduced virulence factor expression in Escherichia coli
- PMID: 39843645
- PMCID: PMC11721078
- DOI: 10.1038/s44259-024-00050-7
YgiV promoter mutations cause resistance to cystobactamids and reduced virulence factor expression in Escherichia coli
Abstract
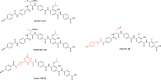
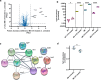
Antimicrobial resistance is one of the major health threats of the modern world. Thus, new structural classes of antimicrobial compounds are needed in order to overcome existing resistance. Cystobactamids represent one such new compound class that inhibit the well-established target bacterial type II topoisomerases while exhibiting superior antibacterial and resistance-breaking properties. Understanding potential mechanisms of emerging resistances is crucial in the development of novel antibiotics as they directly impact the future therapeutic application and market success. Therefore, the frequency and molecular basis of cystobactamid resistance in Escherichia coli was analyzed. High-level resistant E. coli mutants were selected and found to harbor single nucleotide polymorphisms in the promotor region of the ygiV gene, causing an upregulation of the respective protein. These stable mutations are contrary to what was observed as a resistance genotype in the structurally related albicidins, where ygiV gene amplifications were identified as causing resistance. Overexpression of YgiV in the mutants was additionally amplified upon cystobactamid exposition, showing further adaptation to this compound class under treatment. YgiV binds cystobactamids with high binding affinity, thereby preventing their interaction with the antimicrobial targets topoisomerase IV and DNA gyrase. In addition, we observed a substantial impact of YgiV on in vitro gyrase activity by leading to increased DNA cleavage and concurrent reduction in the efficacy of cystobactamids in inhibiting gyrase supercoiling activity. Furthermore, we identified co-upregulation of membrane-modifying proteins, such as EptC, and the transcriptional regulator QseB. This presumably contributes to the observed reduced motility and fimbrial protein expression in resistant mutants, resulting in a reduced expression of virulence factors and potentially pathogenicity, associated with ygiV promotor mutations.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- O´Neill, J. Tackling drug-resistant infections globally: final report and recommendations. Rev. Antimicrob. Resist.7, 110 (2016).
LinkOut - more resources
Full Text Sources

