ARID1A mutations protect follicular lymphoma from FAS-dependent immune surveillance by reducing RUNX3/ETS1-driven FAS-expression
- PMID: 39843653
- PMCID: PMC12089402
- DOI: 10.1038/s41418-025-01445-3
ARID1A mutations protect follicular lymphoma from FAS-dependent immune surveillance by reducing RUNX3/ETS1-driven FAS-expression
Abstract
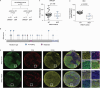
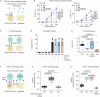
The cell death receptor FAS and its ligand (FASLG) play crucial roles in the selection of B cells during the germinal center (GC) reaction. Failure to eliminate potentially harmful B cells via FAS can lead to lymphoproliferation and the development of B cell malignancies. The classic form of follicular lymphoma (FL) is a prototypic GC-derived B cell malignancy, characterized by the t(14;18)(q32;q21)IGH::BCL2 translocation and overexpression of antiapoptotic BCL2. Additional alterations were shown to be clinically relevant, including mutations in ARID1A. ARID1A is part of the SWI/SNF nucleosome remodeling complex that regulates DNA accessibility ("openness"). However, the mechanism how ARID1A mutations contribute to FL pathogenesis remains unclear. We analyzed 151 FL biopsies of patients with advanced-stage disease at initial diagnosis and found that ARID1A mutations were recurrent and mainly disruptive, with an overall frequency of 18%. Additionally, we observed that ARID1A mutant FL showed significantly lower FAS protein expression in the FL tumor cell population. Functional experiments in BCL2-translocated lymphoma cells demonstrated that ARID1A is directly involved in the regulation of FAS, and ARID1A loss leads to decreased FAS protein and gene expression. However, ARID1A loss did not affect FAS promotor openness. Instead, we identified and experimentally validated a previously unknown co-transcriptional complex consisting of RUNX3 and ETS1 that regulates FAS expression, and ARID1A loss leads to reduced RUNX3 promotor openness and gene expression. The reduced FAS levels induced by ARID1A loss rendered lymphoma cells resistant to both soluble and T cell membrane-anchored FASLG-induced apoptosis, and significantly diminished CAR T cell killing in functional experiments. In summary, we have identified a functionally and clinically relevant mechanism how FL cells can escape FAS-dependent immune surveillance, which may also impact the efficacy of T cell-based therapies, including CAR T cells.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: MSub receives industry research support from Amgen, BMS/Celgene, Gilead, Janssen, Miltenyi Biotec, Morphosys, Novartis, Roche, Seattle Genetics and Takeda; she serves as a consultant/advisor to AvenCell, CDR-Life, Ichnos Sciences, Incyte Biosciences, Janssen, Miltenyi Biotec, Molecular Partners, Novartis, Pfizer and Takeda and serves on the speakers’ bureau at Amgen, AstraZeneca, BMS/Celgene, Gilead, GSK, Janssen, Novartis, Pfizer, Roche and Takeda. OW receives industry research support from Roche and Incyte; he serves as a consultant/advisor to Beigene, Roche, Incyte and Epizyme. All other authors declare no relevant competing interests. Ethics: The research was conducted in compliance with the relevant guidelines and regulations, as outlined in the Guidelines for Safeguarding Good Research Practice, Code of Conduct (German Research Foundation, revised version 1.1). Informed consent was obtained from all patients participating in clinical trials, including consent for the use of biopsy material for research purposes as previously described [11, 20]. Approval for molecular analyzes on human material was obtained from institutional and local ethics committees (LMU #223-14, LMU #276-14, LMU #056/00, and LMU #539-15 fed).
Figures



References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–57. - PubMed
-
- Yoshino T, Kondo E, Cao L, Takahashi K, Hayashi K, Nomura S, et al. Inverse expression of bcl-2 protein and Fas antigen in lymphoblasts in peripheral lymph nodes and activated peripheral blood T and B lymphocytes. Blood. 1994;83:1856–61. - PubMed
MeSH terms
Substances
Grants and funding
- DFG 278529602 - SFB1243 (A11, A12)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- WE 4679/2-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- DFG 278529602 - SFB1243 (A14)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- DFG 278529602 - SFB1243 (A01)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- DFG 278529602 - SFB 1243 (A10)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 2018.087.1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 338/1 2021 - 452881907/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 338/1 2021 - 451580403/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 2022.093.1/Wilhelm Sander-Stiftung (Wilhelm Sander Foundation)
- 2021_EKES.13/Else Kröner-Fresenius-Stiftung (Else Kroner-Fresenius Foundation)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

