Immune evasion through mitochondrial transfer in the tumour microenvironment
- PMID: 39843734
- PMCID: PMC11798832
- DOI: 10.1038/s41586-024-08439-0
Immune evasion through mitochondrial transfer in the tumour microenvironment
Erratum in
-
Publisher Correction: Immune evasion through mitochondrial transfer in the tumour microenvironment.Nature. 2025 Mar;639(8053):E5. doi: 10.1038/s41586-025-08764-y. Nature. 2025. PMID: 39948434 Free PMC article. No abstract available.
-
Author Correction: Immune evasion through mitochondrial transfer in the tumour microenvironment.Nature. 2025 Aug;644(8076):E33. doi: 10.1038/s41586-025-09408-x. Nature. 2025. PMID: 40715527 Free PMC article. No abstract available.
Abstract
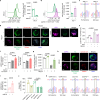
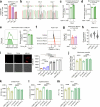
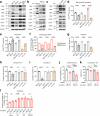
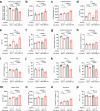
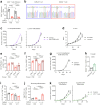
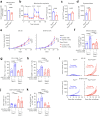
Cancer cells in the tumour microenvironment use various mechanisms to evade the immune system, particularly T cell attack1. For example, metabolic reprogramming in the tumour microenvironment and mitochondrial dysfunction in tumour-infiltrating lymphocytes (TILs) impair antitumour immune responses2-4. However, detailed mechanisms of such processes remain unclear. Here we analyse clinical specimens and identify mitochondrial DNA (mtDNA) mutations in TILs that are shared with cancer cells. Moreover, mitochondria with mtDNA mutations from cancer cells are able to transfer to TILs. Typically, mitochondria in TILs readily undergo mitophagy through reactive oxygen species. However, mitochondria transferred from cancer cells do not undergo mitophagy, which we find is due to mitophagy-inhibitory molecules. These molecules attach to mitochondria and together are transferred to TILs, which results in homoplasmic replacement. T cells that acquire mtDNA mutations from cancer cells exhibit metabolic abnormalities and senescence, with defects in effector functions and memory formation. This in turn leads to impaired antitumour immunity both in vitro and in vivo. Accordingly, the presence of an mtDNA mutation in tumour tissue is a poor prognostic factor for immune checkpoint inhibitors in patients with melanoma or non-small-cell lung cancer. These findings reveal a previously unknown mechanism of cancer immune evasion through mitochondrial transfer and can contribute to the development of future cancer immunotherapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T. Inozume. received honoraria from Ono Pharmaceutical, Bristol-Myers Squibb and MSD outside this study. S.S. received honoraria from AstraZeneca outside this study. Yasuhiro Nakamura. received honoraria from Ono Pharmaceutical and Novartis Pharma outside this study. E.I. received grants from Janssen Pharmaceutical, Bristol-Myers Squibb, Takeda Pharmaceutical, Pfizer Japan and Ono Pharmaceutical; honoraria from AstraZeneca, Novartis Pharma, Janssen Pharmaceutical, Boehringer Ingelheim, Chugai Pharmaceutical, Eli Lilly Japan and Pfizer Japan outside this study. H.H. received research grants from AstraZeneca, Astellas Pharma, MSD, Ono Pharmaceutical, Nippon Boehringer Ingelheim, Novartis Pharma, Pfizer Japan, Bristol-Myers Squibb, Eli Lilly Japan, Chugai Pharmaceutical, Daiichi Sankyo, Merck Serono, Merck Biopharma, Takeda Pharmaceutical, Taiho Pharmaceutical, SymBio Pharmaceuticals, AbbVie, inVentiv Health Japan, ICON Japan, Gritstone Oncology, Parexel International, Kissei Pharmaceutical, EPS, Syneos Health, Pfizer R&D Japan, A2 Healthcare, Quintiles/IQVIA Services Japan, EP-CRSU, Linical, Eisai, CMIC Shift Zero, Kyowa Hakko Kirin, Bayer Yakuhin, EPS International and Otsuka Pharmaceutical; honoraria from Amgen, AstraZeneca, Boehringer Ingelheim Japan, Bristol-Myers Squibb, Chugai Pharmaceutical, Daiichi Sankyo, Eli Lilly Japan, Janssen Pharmaceutical, Kyorin Pharmaceutical, Merck Biopharma, MSD, Novartis Pharmaceuticals, Ono Pharmaceutical, Taiho Pharmaceutical and Takeda Pharmaceutical; consulting fees from AstraZeneca, Boehringer Ingelheim Japan, Bristol-Myers Squibb, Chugai Pharmaceutical, Eli Lilly Japan, Guardant Health, Pfizer Japan, Shanghai Haihe Biopharma, Takeda Pharmaceutical and Merck Biopharma outside this study. Y. Togashi received research grants from KOTAI Biotechnologies, Daiichi-Sankyo, Ono Pharmaceutical, Bristol-Myers Squibb and KORTUC, and honoraria from Ono Pharmaceutical, Bristol-Myers Squibb, AstraZeneca, Chugai Pharmaceutical and MSD outside this study. All other authors declare that they have no competing financial interests.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

