Complete human recombination maps
- PMID: 39843742
- PMCID: PMC11922761
- DOI: 10.1038/s41586-024-08450-5
Complete human recombination maps
Abstract
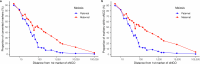
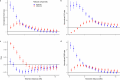
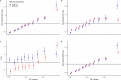
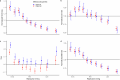
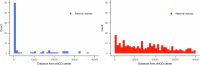
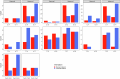
Human recombination maps are a valuable resource for association and linkage studies and crucial for many inferences of population history and natural selection. Existing maps1-5 are based solely on cross-over (CO) recombination, omitting non-cross-overs (NCOs)-the more common form of recombination6-owing to the difficulty in detecting them. Using whole-genome sequence data in families, we estimate the number of NCOs transmitted from parent to offspring and derive complete, sex-specific recombination maps including both NCOs and COs. Mothers have fewer but longer NCOs than fathers, and oocytes accumulate NCOs in a non-regulated fashion with maternal age. Recombination, primarily NCO, is responsible for 1.8% (95% confidence interval: 1.3-2.3) and 11.3% (95% confidence interval: 9.0-13.6) of paternal and maternal de novo mutations, respectively, and may drive the increase in de novo mutations with maternal age. NCOs are substantially more prominent than COs in centromeres, possibly to avoid large-scale genomic changes that may cause aneuploidy. Our results demonstrate that NCOs highlight to a much greater extent than COs the differences in the meiotic process between the sexes, in which maternal NCOs may reflect the safeguarding of oocytes from infancy until ovulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors are employees of deCODE genetics/Amgen.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources

