Antimicrobial peptide glatiramer acetate targets Pseudomonas aeruginosa lipopolysaccharides to breach membranes without altering lipopolysaccharide modification
- PMID: 39843948
- PMCID: PMC11702655
- DOI: 10.1038/s44259-024-00022-x
Antimicrobial peptide glatiramer acetate targets Pseudomonas aeruginosa lipopolysaccharides to breach membranes without altering lipopolysaccharide modification
Abstract
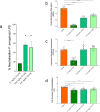
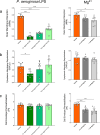
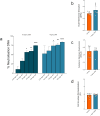
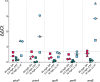
Antimicrobial peptides (AMPs) are key components of innate immunity across all domains of life. Natural and synthetic AMPs are receiving renewed attention in efforts to combat the antimicrobial resistance (AMR) crisis and the loss of antibiotic efficacy. The gram-negative pathogen Pseudomonas aeruginosa is one of the most concerning infecting bacteria in AMR, particularly in people with cystic fibrosis (CF) where respiratory infections are difficult to eradicate and associated with increased morbidity and mortality. Cationic AMPs exploit the negatively charged lipopolysaccharides (LPS) on P. aeruginosa to bind and disrupt bacterial membrane(s), causing lethal damage. P. aeruginosa modifies its LPS to evade AMP killing. Free-LPS is also a component of CF sputum and feeds pro-inflammatory cycles. Glatiramer acetate (GA) is a random peptide co-polymer-of glycine, lysine, alanine, tyrosine-used as a drug in treatment of multiple sclerosis (MS); we have previously shown GA to be an AMP which synergises with tobramycin against CF P. aeruginosa, functioning via bacterial membrane disruption. Here, we demonstrate GA's direct binding and sequestration/neutralisation of P. aeruginosa LPS, in keeping with GA's ability to disrupt the outer membrane. At CF-relevant LPS concentrations, however, membrane disruption by GA was not strongly inhibited. Furthermore, exposure to GA did not result in increased Lipid A modification of LPS or in increased gene expression of systems involved in AMP sensing and LPS modification. Therefore, despite the electrostatic targeting of LPS by GA as part of its activity, P. aeruginosa does not demonstrate LPS modification in its defence.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The Authors declare the following competing interests; Cycle Pharmaceuticals provided Glatiramer Acetate as an in-kind contribution to the CF Trust grant for this work. GA is under license at the University of Aarhus, at which T.V.-J. is employed.
Figures



References
-
- O’Neill, J. Tackling drug-resistant infections globally: final report and recommendations: the review on antimicrobial resistance. 1–35 https://amr-review.org/sites/default/files/160525_Final%20paper_with%20c... (2019).
Grants and funding
LinkOut - more resources
Full Text Sources

