Unravelling genomic drivers of speciation in Musa through genome assemblies of wild banana ancestors
- PMID: 39843949
- PMCID: PMC11754795
- DOI: 10.1038/s41467-025-56329-4
Unravelling genomic drivers of speciation in Musa through genome assemblies of wild banana ancestors
Abstract
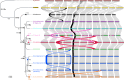
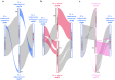
Hybridization between wild Musa species and subspecies from Southeast Asia is at the origin of cultivated bananas. The genomes of these cultivars are complex mosaics involving nine genetic groups, including two previously unknown contributors. This study provides continuous genome assemblies for six wild genetic groups, one of which represents one of the unknown ancestor, identified as M.acuminata ssp. halabanensis. The second unknown ancestor partially present in a seventh assembly appears related to M. a. ssp. zebrina. These assemblies provide key resources for banana genetics and for improving cultivar assemblies, including that of the emblematic triploid Cavendish. Comparative and phylogenetic analyses reveal an ongoing speciation process within Musa, characterised by large chromosome rearrangements and centromere differentiation through the integration of different types of repeated sequences, including rDNA tandem repeats. This speciation process may have been favoured by reproductive isolation related to the particular context of climate and land connectivity fluctuations in the Southeast Asian region.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Interspecific introgression patterns reveal the origins of worldwide cultivated bananas in New Guinea.Plant J. 2023 Feb;113(4):802-818. doi: 10.1111/tpj.16086. Epub 2023 Jan 18. Plant J. 2023. PMID: 36575919
-
Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana.Plant J. 2020 Jun;102(5):1008-1025. doi: 10.1111/tpj.14683. Epub 2020 Feb 28. Plant J. 2020. PMID: 31930580 Free PMC article.
-
Origins and domestication of cultivated banana inferred from chloroplast and nuclear genes.PLoS One. 2013 Nov 18;8(11):e80502. doi: 10.1371/journal.pone.0080502. eCollection 2013. PLoS One. 2013. PMID: 24260405 Free PMC article.
-
Domestication, genomics and the future for banana.Ann Bot. 2007 Nov;100(5):1073-84. doi: 10.1093/aob/mcm191. Epub 2007 Aug 31. Ann Bot. 2007. PMID: 17766312 Free PMC article. Review.
-
Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana.Cytogenet Genome Res. 2005;109(1-3):27-33. doi: 10.1159/000082378. Cytogenet Genome Res. 2005. PMID: 15753555 Review.
References
-
- Woodruff, D. S. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodivers. Conserv.19, 919–941 (2010).
-
- Häkkinen, M. Reappraisal of sectional taxonomy in Musa (Musaceae). TAXON62, 809–813 (2013).
-
- Simmonds, N. W. The Evolution of the Bananas. (Longmans, London, 1962).
-
- Simmonds, N. W. & Shepherd, K. The taxonomy and origins of the cultivated bananas. J. Linn. Soc. Lond. Bot.55, 302–312 (1955).
MeSH terms
LinkOut - more resources
Full Text Sources

