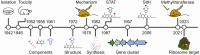
History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome
- PMID: 39843956
- PMCID: PMC11702664
- DOI: 10.1038/s44259-023-00020-5
History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome
Abstract
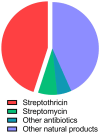
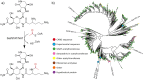
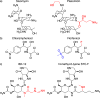
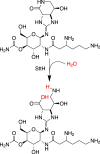
The streptothricin antibiotics were among the first antibiotics to be discovered from the environment and remain some of the most recovered antimicrobials in natural product screens. Increasing rates of antibiotic resistance and recognition that streptothricin antibiotics may play a role in countering so-called super-bugs has led to the re-evaluation of their clinical potential. Here we will review the current state of knowledge of streptothricins and their resistance in bacteria, with a focus on the potential for new resistance mechanisms and determinants to emerge in the context of potential widespread clinical adoption of this antibiotic class.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- O’Neill, J. Tackling drug-resistant infections globally: final report and recommendations. https://wellcomecollection.org/works/thvwsuba (2016).
-
- Talkington, K., Shore, C. & Kothari, P. A Scientific Roadmap for Antibiotic Discovery. https://www.pewtrusts.org/-/media/assets/2016/05/ascientificroadmapforan... (2016).
Publication types
LinkOut - more resources
Full Text Sources

