Preserving information while respecting privacy through an information theoretic framework for synthetic health data generation
- PMID: 39843957
- PMCID: PMC11754479
- DOI: 10.1038/s41746-025-01431-6
Preserving information while respecting privacy through an information theoretic framework for synthetic health data generation
Abstract
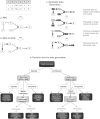
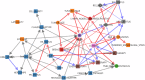
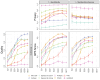
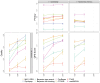
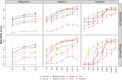
Generating synthetic data from medical records is a complex task intensified by patient privacy concerns. In recent years, multiple approaches have been reported for the generation of synthetic data, however, limited attention was given to jointly evaluate the quality and the privacy of the generated data. The quality and privacy of synthetic data stem from multivariate associations across variables, which cannot be assessed by comparing univariate distributions with the original data. Here, we introduce a novel algorithm (MIIC-SDG) for generating synthetic data from electronic records based on a multivariate information framework and Bayesian network theory. We also propose a new metric to quantitatively assess the trade-off between the Quality and Privacy Scores (QPS) of synthetic data generation methods. The performance of MIIC-SDG is demonstrated on different clinical datasets and favorably compares with state-of-the-art synthetic data generation methods, based on the QPS trade-off between several quality and privacy metrics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests
Figures



References
-
- Samarati, P. & Sweeney, L. Protecting Privacy When Disclosing Information: k-Anonymity and Its Enforcement Through Generalization and Suppression.https://www.semanticscholar.org (1998).
-
- Machanavajjhala, A., Kifer, D., Gehrke, J. & Venkitasubramaniam, M. L-diversity: Privacy beyond k-anonymity. ACM Trans. Knowl. Discov. Data1, 3-es (2007). - DOI
-
- Li, N., Li, T. & Venkatasubramanian, S. t-closeness: Privacy beyond k-anonymity and l-diversity. In 2007 IEEE 23rd International Conference on Data Engineering 106–115 (IEEE, 2007).
-
- Hernandez, M., Epelde, G., Alberdi, A., Cilla, R. & Rankin, D. Synthetic data generation for tabular health records: a systematic review. Neurocomputing493, 28–45 (2018).
LinkOut - more resources
Full Text Sources

