AaMYB61-like and AabHLH137 jointly regulate anthocyanin biosynthesis in Actinidia arguta
- PMID: 39844047
- PMCID: PMC11753137
- DOI: 10.1186/s12870-025-06109-5
AaMYB61-like and AabHLH137 jointly regulate anthocyanin biosynthesis in Actinidia arguta
Abstract
Background: Red Actinidia arguta has recently become highly popular because of its red appearance resulting from anthocyanin accumulation, and has gradually become an important breeding direction. However, regulators involved in anthocyanin biosynthesis have not been fully characterized in A. arguta.
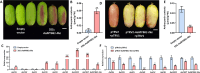
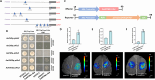
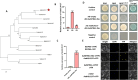
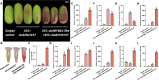
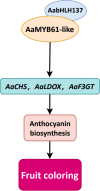
Results: Here, we demonstrated that a key R2R3-MYB transcription factor, AaMYB61-like, plays a crucial role in A. arguta anthocyanin biosynthesis. The RT-qPCR results revealed that transient overexpression of AaMYB61-like in A. arguta fruit at 90-100 DAFB significantly promoted anthocyanin biosynthesis, as did the gene expression levels of AaCHS, AaCHI, AaF3H, AaLDOX, and AaF3GT, whereas the result of VIGS revealed the opposite results in A. arguta fruit at 105-115 DAFB. A transcriptional activation assay indicated that AaMYB61-like exhibited transcriptional activation activity. Y1H and LUC assays revealed that AaMYB61-like activates the promoters of AaCHS, AaLDOX, and AaF3GT. In addition, AabHLH137 was found to be related to fruit color from the transcriptome data. We demonstrated that AaMYB61-like promotes anthocyanin biosynthesis by interacting with AabHLH137 via Y2H, BiFC, and Agrobacterium-mediated co-transformation.
Conclusions: Our study not only reveals the functions of AaMYB61-like and AabHLH137 in anthocyanin regulation, but also broadly enriches color regulation theory, establishing a foundation for clarifying the molecular mechanism of fruit coloration in kiwifruit.
Keywords: Actinidia arguta; AaMYB61-like; AabHLH137; Anthocyanin biosynthesis; Transcription regulation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study did not include human or animal subjects. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Li Y, Qi X, Cui W, Lin M, Qiao C, Zhong Y, et al. Restraint of Bagging on Fruit Skin Coloration in on-Tree Kiwifruit (Actinidia arguta). J Plant Growth Regul. 2021;40:603–16. - DOI
MeSH terms
Substances
Grants and funding
- 32202435/National Natural Science Foundation of China
- 2022YFD1200503/National Key Research and Development Program of China
- 2022YFD1600700/National Key Research and Development Program of China
- 2024(026)/Major Scientific and Technological Special Project of Guizhou Province
- HARS-22-09-S/Technical System of Bulk Fruit Industry in Henan Province
LinkOut - more resources
Full Text Sources

