Single-cell profiling of SLC family transporters: uncovering the role of SLC7A1 in osteosarcoma
- PMID: 39844299
- PMCID: PMC11752724
- DOI: 10.1186/s12967-025-06086-1
Single-cell profiling of SLC family transporters: uncovering the role of SLC7A1 in osteosarcoma
Abstract
Background: Osteosarcoma is the most common malignant bone tumor in children and adolescents, characterized by high disability and mortality rates. Over the past three decades, therapeutic outcomes have plateaued, underscoring the critical need for innovative therapeutic targets. Solute carrier (SLC) family transporters have been implicated in the malignant progression of a variety of tumors, however, their specific role in osteosarcoma remains poorly understood.
Methods: The single-cell sequencing data from GSE152048 and GSE162454, along with RNA-seq from the TARGET and GSE21257 cohorts, were utilized for the analysis in this study. LASSO regression analysis was conducted to identify prognostic genes and construct an SLC-related prognostic signature. Survival analysis and ROC analysis evaluated the validity of the prognostic signature. The ESTIMATE and CIBERSORT Packages were utilized to assess the immune infiltration status. Pseudotime and CellChat analyses were performed to investigate the relationship between SLC7A1, malignant phenotypes, and the immune microenvironment. CCK8 assays, EdU staining, colony formation assays, Transwell assays, and co-culture systems were used to assess the effects of SLC7A1 on cell proliferation, metastasis, and macrophage polarization. Finally, virtual docking identified potential drugs targeting SLC7A1.
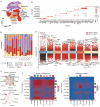
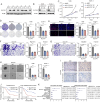
Results: SLCs displayed distinct expression patterns across various cell types within the osteosarcoma microenvironment, with myeloid cells exhibiting a preference for amino acid uptake. A prognostic model comprising nine genes was constructed via LASSO regression, with SLC7A1 showing the highest hazard ratio. Multiple analytical algorithms indicated that SLCs were associated with immune cell infiltration and immune checkpoint gene expression. Single-cell analysis indicated that SLC7A1 was predominantly expressed in osteosarcoma cells and correlated with various malignant tumor characteristics. SLC7A1 also regulate interactions between tumor cells and macrophages, as well as modulate macrophage function through multiple pathways. In vitro assays and survival analysis demonstrated that inhibition of SLC7A1 suppressed the malignant phenotype of osteosarcoma cells, with SLC7A1 expression correlating with poor prognosis. Co-culture models confirmed the involvement of SLC7A1 in macrophage polarization. Finally, virtual screening and CETSA identified Cepharanthine as potential inhibitors of SLC7A1.
Conclusion: SLC-related prognostic signatures can be utilized for the prognostic evaluation of osteosarcoma. Pharmacological inhibition of SLC7A1 may be a feasible therapeutic approach for osteosarcoma.
Keywords: Osteosarcoma; SLC7A1; Solute carrier (SLC) family transporters; TAMs; Tumor immune microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Human sample collection for this study was conducted in accordance with the Declaration of Helsinki, and was approved by the Research Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University, with the reference number [2021]755. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures








References
-
- Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncology: Official J Am Soc Clin Oncol. 2018;36(2):188–93. - PubMed
-
- Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Reviews Clin Oncol. 2021;18(10):609–24. - PubMed
-
- Ritter J, Bielack SS, Osteosarcoma. Annals of Oncology. Official J Eur Soc Med Oncol. 2010;21(Suppl 7):vii320–5. - PubMed
-
- Meltzer PS, Helman LJ. New Horizons in the Treatment of Osteosarcoma. N Engl J Med. 2021;385(22):2066–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

