A novel approach for detecting Salmonella enterica strains frequently attributed to human illness-development and validation of the highly pathogenic Salmonella (HPS) multiplex PCR assay
- PMID: 39845055
- PMCID: PMC11752890
- DOI: 10.3389/fmicb.2024.1504621
A novel approach for detecting Salmonella enterica strains frequently attributed to human illness-development and validation of the highly pathogenic Salmonella (HPS) multiplex PCR assay
Abstract
Introduction: Non-typhoidal Salmonella enterica (NTS) are leading bacterial agents of foodborne illnesses and a global concern for human health. While there are over 2,600 different serovars of NTS, epidemiological data suggests that certain serovars are better at causing disease than others, resulting in the majority of reported human illnesses in the United States. To improve food safety, there is a need to rapidly detect these more pathogenic serovars to facilitate their removal from the food supply.
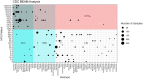
Methods: Addressing this need, we conducted a comparative analysis of 23 closed Salmonella genomic sequences of five serotypes. The analysis pinpointed eight genes (sseK2, sseK3, gtgA/gogA, avrA, lpfB, SspH2, spvD, and invA) that in combination, identify 7 of the 10 leading Salmonella serovars attributed to human illnesses in the US each year (i.e., Serovars of Concern or SoC). A multiplex PCR assay was developed to detect the presence of these genes, with strains amplifying five or more targets designated Highly Pathogenic Salmonella, or HPS. The utility of the resulting HPS assay for identifying SoC was examined in silico, using BLAST to determine the distribution of gene targets among closed Salmonella genome sequences in GenBank (n = 2,192 representing 148 serotypes) and by assaying 1,303 Salmonella (69 serotypes), isolated from FSIS regulatory samples.
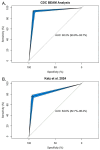
Results and discussion: Comparison of serotypes identified by the assay as HPS, with those identified as SoC, produced an Area Under the Curve (AUC) of 92.2% with a specificity of 96% and a positive predictive value of 97.4%, indicating the HPS assay has strong ability to identify SoC. The data presented lay the groundwork for development of rapid commercial assays for the detection of SoC.
Keywords: fimbriae; highly pathogenic Salmonella (HPS); multiplex PCR; non-typhoidal Salmonella enterica; secreted effector; serovars of concern (SoC).
Copyright © 2025 Harhay, Brader, Katz, Harhay, Bono, Bosilevac and Wheeler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- BLAST: Basic Local Alignment Search Tool (n.d.). Available at: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed September 26, 2024).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

