Insights into microbial compositions of the respiratory tract of neonatal dairy calves in a longitudinal probiotic trial through 16S rRNA sequencing
- PMID: 39845057
- PMCID: PMC11751226
- DOI: 10.3389/fmicb.2024.1499531
Insights into microbial compositions of the respiratory tract of neonatal dairy calves in a longitudinal probiotic trial through 16S rRNA sequencing
Abstract
Introduction: Probiotics are a promising intervention for modulating the microbiome and the immune system, promoting health benefits in cattle. While studies have characterized the calf lung bacterial profile with and without oral probiotics, simultaneous probiotic effects on the bacterial populations of multiple sites along the respiratory tract have not been characterized.
Methods: This study utilized the same pre-weaning diary calf group from our previous studies to characterize the bacterial populations present in the nostril and tonsil across control and treatment groups and nine sampling time points. DNA was exacted from the nostril and tonsil swabs and lung lavage fluids, and 16S ribosomal RNA gene hypervariable regions 1-3 were subsequently sequenced.
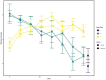
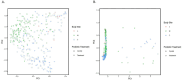
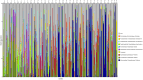
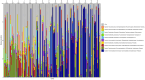
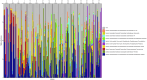
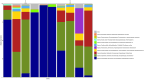
Results: Temporal variation in alpha bacterial diversity within the nostril, tonsil, and lung samples was observed, indicating distinct bacterial compositions among sampling time points. Oral probiotic treatment did not change alpha diversity in any respiratory tissue, however, spatial variability in bacterial taxa composition was observed among the three respiratory tract regions. While the majority of differentially abundant taxa in probiotic treated calves were unique to their anatomical location, a few were common to two anatomical locations and one Finegoldia amplicon sequence variant was differentially abundant in all three anatomical locations.
Discussion: In conclusion, these findings contribute to the understanding of the dynamic nature of bacterial diversity and the potential effects of probiotics within the bovine respiratory tract and provides insight for future studies of probiotics on animal health, disease prevention, and management.
Keywords: 16S rRNA; dairy cattle; microbiome; probiotic; respiratory.
Copyright © 2025 Tan, Eicher, Kritchevsky, Bryan, Dickey, Chitko-McKown and McDaneld.
Conflict of interest statement
KB was employed by Chr. Hansen, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Adjei-Fremah S., Ekwemalor K., Asiamah E. K., Ismail H., Ibrahim S., Worku M. (2018). Effect of probiotic supplementation on growth and global gene expression in dairy cows. J. Appl. Anim. Res. 46, 257–263. doi: 10.1080/09712119.2017.1292913 - DOI
-
- Alayande K. A., Aiyegoro O. A., Ateba C. N. (2020). Probiotics in animal husbandry: applicability and associated risk factors. Sustain. For. 12:1087. doi: 10.3390/su12031087 - DOI
-
- Al-Shawi S. G., Dang D. S., Yousif A. Y., Al-Younis Z. K., Najm T. A., Matarneh S. K. (2020). The potential use of probiotics to improve animal health, efficiency, and meat quality: a review. Agriculture 10:452. doi: 10.3390/agriculture10100452 - DOI
LinkOut - more resources
Full Text Sources

