Miniature-inverted-repeat transposable elements contribute to phenotypic variation regulation of rice induced by space environment
- PMID: 39845491
- PMCID: PMC11751223
- DOI: 10.3389/fpls.2024.1446383
Miniature-inverted-repeat transposable elements contribute to phenotypic variation regulation of rice induced by space environment
Abstract
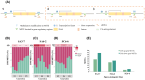
Introduction: Rice samples exposed to the space environment have generated diverse phenotypic variations. Miniature-inverted-repeat transposable elements (MITEs), often found adjacent to genes, play a significant role in regulating the plant genome. Herein, the contribution of MITEs in regulating space-mutagenic phenotypes was explored.
Methods: The space-mutagenic phenotype changes in the F3 to F5 generations of three space-mutagenic lines from the rice varieties Dongnong423 (DN423) and Dongnong (DN416) were meticulously traced. Rice leaves samples at the heading stage from three space-mutagenic lines were subjected to high coverage whole-genome bisulfite sequencing and whole-genome sequencing. These analyses were conducted to investigate the effects of MITEs related epigenetic and genetic variations on space-mutagenic phenotypes.
Results and discussion: Studies have indicated that MITEs within gene regulatory regions might contribute to the formation and differentiation of space-mutagenic phenotypes. The space environment has been shown to induce the transposable elements insertion polymorphisms of MITEs (MITEs-TIPs), with a notable preference for insertion near genes involved in stress response and phenotype regulation. The space-induced MITEs-TIPs contributed to the formation of space-mutagenic phenotype by modulating the expression of gene near the insertion site. This study underscored the pivotal role of MITEs in modulating plant phenotypic variation induced by the space environment, as well as the transgenerational stability of these phenotypic variants.
Keywords: TEs; environment stress; mites; phenotype variation; rice; space environment; space mutagenesis.
Copyright © 2025 Chen, Yang, Zhang and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

