Alternative splicing of modulatory immune receptors in T lymphocytes: a newly identified and targetable mechanism for anticancer immunotherapy
- PMID: 39845971
- PMCID: PMC11752881
- DOI: 10.3389/fimmu.2024.1490035
Alternative splicing of modulatory immune receptors in T lymphocytes: a newly identified and targetable mechanism for anticancer immunotherapy
Abstract
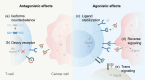
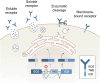
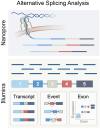
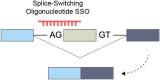
Alternative splicing (AS) is a mechanism that generates translational diversity within a genome. Equally important is the dynamic adaptability of the splicing machinery, which can give preference to one isoform over others encoded by a single gene. These isoform preferences change in response to the cell's state and function. Particularly significant is the impact of physiological alternative splicing in T lymphocytes, where specific isoforms can enhance or reduce the cells' reactivity to stimuli. This process makes splicing isoforms defining features of cell states, exemplified by CD45 splice isoforms, which characterize the transition from naïve to memory states. Two developments have accelerated the use of AS dynamics for therapeutic interventions: advancements in long-read RNA sequencing and progress in nucleic acid chemical modifications. Improved oligonucleotide stability has enabled their use in directing splicing to specific sites or modifying sequences to enhance or silence particular splicing events. This review highlights immune regulatory splicing patterns with potential significance for enhancing anticancer immunotherapy.
Keywords: T lymphocytes; alternative splicing; cancer; immune receptors; immunotherapy.
Copyright © 2025 Tzaban, Stern, Zisman, Eisenberg, Klein, Frankenburg and Lotem.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


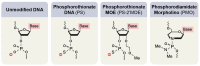
References
-
- Holmes ME, Hertel KJ. Interdependent regulation of alternative splicing by SR and hnRNP proteins. bioRxiv. (2024). doi: 10.1101/2024.08.19.608666 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

