Ultra pure high molecular weight DNA from soil for Nanopore shotgun metagenomics and metabarcoding sequencing
- PMID: 39846015
- PMCID: PMC11751509
- DOI: 10.1016/j.mex.2024.103134
Ultra pure high molecular weight DNA from soil for Nanopore shotgun metagenomics and metabarcoding sequencing
Abstract
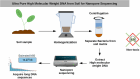
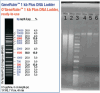
Soil microbes are among the most abundant and diverse organisms on Earth but remain poorly characterized. New technologies have made possible to sequence the DNA of uncultivated microorganisms in soil and other complex ecosystems. Genome assembly is crucial for understanding their functional potential. Nanopore sequencing technologies allow to sequence long DNA fragments, optimizing production of metagenome-assembled genomes compared to short-read technology. Extracting DNA with a very high purity and high molecular weight is key to get the most out of this long read technologies. Here we present two extraction protocols to get DNA with high purity. First protocol is optimized to reach DNA quality suiting Nanopore shotgun metagenomics. It uses a non-toxic centrifugation gradient to separate bacterial cells from soil to extract DNA directly on cells. The median length of the acquired DNA sequences (N50) was 3 to 7 times greater than previously published in the literature, achieving an N50 of ∼14 kb. The other, a modification of a commercially available MP Biomedical DNA extraction kit, yielded high-purity DNA for full-length 16S Oxford Nanopore metabarcoding, with an N50 of ∼8 kb. The MP-based protocol achieves higher yields of ultra-pure DNA compared to the Nycodenz protocol, at the expense of shorter fragment lengths.
Keywords: Bacterial cells; Community DNA; Long fragments; MP kit; Metabarcoding; Metagenomics - Oxford nanopore nanopore sequencing; Nycodenz; Solvent free; Ultra Pure HMW DNA extraction and purification from soil for Nanopore sequencing.
© 2024 Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Lindahl V., Bakken L.R. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiol. Ecol. Feb. 1995;16(2):135–142. doi: 10.1111/j.1574-6941.1995.tb00277.x. - DOI
-
- Dequiedt S., et al. Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management: biogeography of soil microbial biomass. Global Ecol. Biogeogr. Jul. 2011;20(4):641–652. doi: 10.1111/j.1466-8238.2010.00628.x. - DOI
LinkOut - more resources
Full Text Sources

