CD34hi subset of synovial fibroblasts contributes to fibrotic phenotype of human knee osteoarthritis
- PMID: 39846253
- PMCID: PMC11790023
- DOI: 10.1172/jci.insight.183690
CD34hi subset of synovial fibroblasts contributes to fibrotic phenotype of human knee osteoarthritis
Abstract
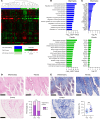
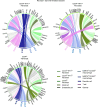
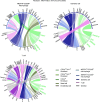
Osteoarthritis (OA) shows various clinical manifestations depending on the status of its joint components. We aimed to identify the synovial cell subsets responsible for OA pathophysiology by comprehensive analyses of human synovium samples in single-cell resolution. Two distinct OA synovial tissue groups were classified by gene expression profiles in RNA-Seq: inflammatory and fibrotic. The inflammatory group exhibited high expression of inflammatory cytokines, histologically inflammatory infiltrate, and a more severe pain score. The fibrotic group showed higher expression of fibroblast growth factor (FGFs) and bone morphogenetic proteins (BMPs), showed histologically perivascular fibrosis, and showed a lower pain score. In single-cell RNA-Seq (scRNA-Seq) of synovial cells, MERTKloCD206lo macrophages and CD34hi fibroblasts were associated with the inflammatory and fibrotic groups, respectively. Among the 3 fibroblast subsets, CD34loTHY1lo and CD34loTHY1hi fibroblasts were influenced by synovial immune cells, whereas CD34hi fibroblasts were influenced by mural and endothelial cells. Particularly, in CD34hi fibroblast subsets, CD34hiCD70hi fibroblasts promoted proliferation of Tregs, potentially suppressing synovitis and protecting articular cartilage. Elucidation of the mechanisms underlying the regulation of these synovial cell subsets may lead to novel strategies for OA therapeutics.
Keywords: Bone biology; Inflammation; Osteoarthritis.
Figures





References
-
- Moskowitz RW, et al, eds. Osteoarthritis: Diagnosis and Medical/Surgical Management. 4th Edition. Lippincott Williams & Wilkins; 2007.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

