Peptide Inhibitor Assay for Allocating Functionally Important Accessible Sites Throughout a Protein Chain: Restriction Endonuclease EcoRI as a Model Protein System
- PMID: 39846550
- PMCID: PMC11755562
- DOI: 10.3390/biotech14010001
Peptide Inhibitor Assay for Allocating Functionally Important Accessible Sites Throughout a Protein Chain: Restriction Endonuclease EcoRI as a Model Protein System
Abstract
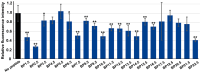
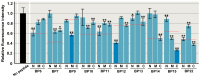
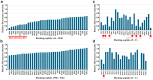
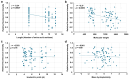
Functionally important amino acid sequences in proteins are often located at multiple sites. Three-dimensional structural analysis and site-directed mutagenesis may be performed to allocate functional sites for understanding structure‒function relationships and for developing novel inhibitory drugs. However, such methods are too demanding to comprehensively cover potential functional sites throughout a protein chain. Here, a peptide inhibitor assay (PIA) was devised to allocate functionally important accessible sites in proteins. This simple method presumes that protein‒ligand interactions, intramolecular interactions, and dimerization interactions can be partially inhibited by high concentrations of competitive "endogenous" peptides of the protein of interest. Focusing on the restriction endonuclease EcoRI as a model protein system, many endogenous peptides (6mer-14mer) were synthesized, covering the entire EcoRI protein chain. Some of them were highly inhibitory, but interestingly, the nine most effective peptides were located outside the active sites, with the exception of one. Relatively long peptides with aromatic residues (F, H, W, and Y) corresponding to secondary structures were generally effective. Because synthetic peptides are flexible enough to change length and amino acid residues, this method may be useful for quickly and comprehensively understanding structure‒function relationships and developing novel drugs or epitopes for neutralizing antibodies.
Keywords: EcoRI; drug design; molecular accessibility; peptide and protein engineering; peptide inhibitor; protein structure and function.
Conflict of interest statement
The author declares no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Inhibiting the dimeric restriction endonuclease EcoRI using interfacial helical peptides.Chem Biol. 1998 Jun;5(6):339-43. doi: 10.1016/s1074-5521(98)90172-7. Chem Biol. 1998. PMID: 9653552
-
Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues.J Mol Biol. 1998 Jun 5;279(2):473-81. doi: 10.1006/jmbi.1998.1803. J Mol Biol. 1998. PMID: 9642051
-
Mutants of the EcoRI endonuclease with promiscuous substrate specificity implicate residues involved in substrate recognition.EMBO J. 1990 Oct;9(10):3369-78. doi: 10.1002/j.1460-2075.1990.tb07538.x. EMBO J. 1990. PMID: 2209548 Free PMC article.
-
How the EcoRI endonuclease recognizes and cleaves DNA.Bioessays. 1992 Jul;14(7):445-54. doi: 10.1002/bies.950140704. Bioessays. 1992. PMID: 1445286 Review.
-
Human follitropin heterodimerization and receptor binding structural motifs: identification and analysis by a combination of synthetic peptide and mutagenesis approaches.Mol Cell Endocrinol. 1996 Dec 20;125(1-2):45-54. doi: 10.1016/s0303-7207(96)03947-0. Mol Cell Endocrinol. 1996. PMID: 9027342 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
