Identification of Proteoforms Related to Nelumbo nucifera Flower Petaloid Through Proteogenomic Strategy
- PMID: 39846635
- PMCID: PMC11755666
- DOI: 10.3390/proteomes13010004
Identification of Proteoforms Related to Nelumbo nucifera Flower Petaloid Through Proteogenomic Strategy
Abstract
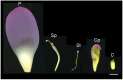
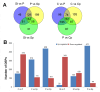
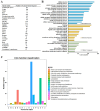
Nelumbo nucifera is an aquatic plant with a high ornamental value due to its flower. Despite the release of several versions of the lotus genome, its annotation remains inefficient, which makes it difficult to obtain a more comprehensive knowledge when -omic studies are applied to understand the different biological processes. Focusing on the petaloid of the lotus flower, we conducted a comparative proteomic analysis among five major floral organs. The proteogenomic strategy was applied to analyze the mass spectrometry data in order to dig out novel proteoforms that are involved in the petaloids of the lotus flower. The results revealed that a total of 4863 proteins corresponding to novel genes were identified, with 227 containing single amino acid variants (SAAVs), and 72 originating from alternative splicing (AS) genes. In addition, a range of post-translational modifications (PTMs) events were also identified in lotus. Through functional annotation and homology analysis with 24 closely related plant species, we identified five candidate proteins associated with floral organ development, which were not identified by ordinary proteomic analysis. This study not only provides new insights into understanding the mechanism of petaloids in lotus but is also helpful in identifying new proteoforms to improve the annotation of the lotus genome.
Keywords: Nelumbo nucifera; new proteoforms; petaloidy; proteogenomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

