RNA Metabolism and the Role of Small RNAs in Regulating Multiple Aspects of RNA Metabolism
- PMID: 39846679
- PMCID: PMC11755482
- DOI: 10.3390/ncrna11010001
RNA Metabolism and the Role of Small RNAs in Regulating Multiple Aspects of RNA Metabolism
Abstract
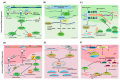
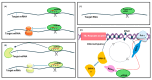
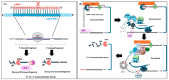
RNA metabolism is focused on RNA molecules and encompasses all the crucial processes an RNA molecule may or will undergo throughout its life cycle. It is an essential cellular process that allows all cells to function effectively. The transcriptomic landscape of a cell is shaped by the processes such as RNA biosynthesis, maturation (RNA processing, folding, and modification), intra- and inter-cellular transport, transcriptional and post-transcriptional regulation, modification, catabolic decay, and retrograde signaling, all of which are interconnected and are essential for cellular RNA homeostasis. In eukaryotes, sRNAs, typically 20-31 nucleotides in length, are a class of ncRNAs found to function as nodes in various gene regulatory networks. sRNAs are known to play significant roles in regulating RNA population at the transcriptional, post-transcriptional, and translational levels. Along with sRNAs, such as miRNAs, siRNAs, and piRNAs, new categories of ncRNAs, i.e., lncRNAs and circRNAs, also contribute to RNA metabolism regulation in eukaryotes. In plants, various genetic screens have demonstrated that sRNA biogenesis mutants, as well as RNA metabolism pathway mutants, exhibit similar growth and development defects, misregulated primary and secondary metabolism, as well as impaired stress response. In addition, sRNAs are both the "products" and the "regulators" in broad RNA metabolism networks; gene regulatory networks involving sRNAs form autoregulatory loops that affect the expression of both sRNA and the respective target. This review examines the interconnected aspects of RNA metabolism with sRNA regulatory pathways in plants. It also explores the potential conservation of these pathways across different kingdoms, particularly in plants and animals. Additionally, the review highlights how cellular RNA homeostasis directly impacts adaptive responses to environmental changes as well as different developmental aspects in plants.
Keywords: PTGS; RNA editing; RNA modification; RNA processing; Retrograde signaling; TGS; Transcription; mRNA; mRNA decay; non-coding RNAs; small RNAs.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

