LncRNA 3222401L13Rik Is Upregulated in Aging Astrocytes and Regulates Neuronal Support Function Through Interaction with Npas3
- PMID: 39846680
- PMCID: PMC11755665
- DOI: 10.3390/ncrna11010002
LncRNA 3222401L13Rik Is Upregulated in Aging Astrocytes and Regulates Neuronal Support Function Through Interaction with Npas3
Abstract
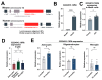
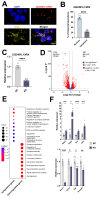
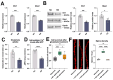
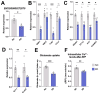
Aging leads to cognitive decline and increased risk of neurodegenerative diseases. While molecular changes in central nervous system (CNS) cells contribute to this decline, the mechanisms are not fully understood. Long non-coding RNAs (lncRNAs) are key regulators of cellular functions. Background/Objectives: The roles of lncRNAs in aging, especially in glial cells, are not well characterized. Methods: We investigated lncRNA expression in non-neuronal cells from aged mice and identified 3222401L13Rik, a previously unstudied lncRNA, as upregulated in astrocytes during aging. Results: Knockdown of 3222401L13Rik in primary astrocytes revealed its critical role in regulating genes for neuronal support and synapse organization, a function conserved in human iPSC-derived astrocytes. A 3222401L13Rik interacts with the transcription factor Neuronal PAS Domain Protein 3 (Npas3), and overexpression of Npas3 rescues deficits in astrocytes lacking 3222401L13Rik. Conclusions: These data suggest that 3222401L13Rik upregulation may help delay age-related cognitive decline.
Keywords: Alzheimer’s disease; aging; astrocytes; brain; lncRNA; neurodegenerative diseases; non-coding RNA; transcriptomics.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
-
Integrative RNA profiling of TBEV-infected neurons and astrocytes reveals potential pathogenic effectors.Comput Struct Biotechnol J. 2022 May 30;20:2759-2777. doi: 10.1016/j.csbj.2022.05.052. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35685361 Free PMC article.
-
Npas3 deficiency impairs cortical astrogenesis and induces autistic-like behaviors.Cell Rep. 2022 Aug 30;40(9):111289. doi: 10.1016/j.celrep.2022.111289. Cell Rep. 2022. PMID: 36044858
-
IL-9-triggered lncRNA Gm13568 regulates Notch1 in astrocytes through interaction with CBP/P300: contribute to the pathogenesis of experimental autoimmune encephalomyelitis.J Neuroinflammation. 2021 May 11;18(1):108. doi: 10.1186/s12974-021-02156-5. J Neuroinflammation. 2021. PMID: 33971906 Free PMC article.
-
Regulatory Roles of Long Non-Coding RNAs in the Central Nervous System and Associated Neurodegenerative Diseases.Front Cell Neurosci. 2017 Jun 30;11:175. doi: 10.3389/fncel.2017.00175. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28713244 Free PMC article. Review.
Cited by
-
Single-Cell Epigenomics Uncovers Heterochromatin Instability and Transcription Factor Dysfunction during Mouse Brain Aging.bioRxiv [Preprint]. 2025 May 20:2025.04.21.649585. doi: 10.1101/2025.04.21.649585. bioRxiv. 2025. PMID: 40475496 Free PMC article. Preprint.
References
-
- Berchtold N.C., Cribbs D.H., Coleman P.D., Rogers J., Head E., Kim R., Beach T., Miller C., Troncoso J., Trojanowski J.Q., et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. USA. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. - DOI - PMC - PubMed
-
- Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Java S., Agis-Balboa R.C., Cota P., Wittnam J., Gogul-Doering A., Opitz L., et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. - DOI - PubMed
LinkOut - more resources
Full Text Sources

