Anti-HIV-1 Effect of the Fluoroquinolone Enoxacin and Modulation of Pro-Viral hsa-miR-132 Processing in CEM-SS Cells
- PMID: 39846686
- PMCID: PMC11755467
- DOI: 10.3390/ncrna11010008
Anti-HIV-1 Effect of the Fluoroquinolone Enoxacin and Modulation of Pro-Viral hsa-miR-132 Processing in CEM-SS Cells
Erratum in
-
Correction: Schlösser et al. Anti-HIV-1 Effect of the Fluoroquinolone Enoxacin and Modulation of Pro-Viral hsa-miR-132 Processing in CEM-SS Cells. Non-Coding RNA 2025, 11, 8.Noncoding RNA. 2025 May 16;11(3):40. doi: 10.3390/ncrna11030040. Noncoding RNA. 2025. PMID: 40407598 Free PMC article.
Abstract
Background: Despite tremendous advances in antiretroviral therapy (ART) against HIV-1 infections, no cure or vaccination is available. Therefore, discovering novel therapeutic strategies remains an urgent need. In that sense, miRNAs and miRNA therapeutics have moved intensively into the focus of recent HIV-1-related investigations. A strong reciprocal interdependence has been demonstrated between HIV-1 infection and changes of the intrinsic cellular miRNA milieu. This interrelationship may direct potential alterations of the host cells' environment beneficial for the virus or its suppression of replication. Whether this tightly balanced and controlled battle can be exploited therapeutically remains to be further addressed. In this context, the fluoroquinolone antibiotic Enoxacin has been demonstrated as a potent modulator of miRNA processing. Here, we test the hypothesis that this applies also to selected HIV-1-related miRNAs.
Methods: We studied the effect of Enoxacin on HIV-1 replication coupled with miRNA qRT-PCR analysis of HIV-1-related miRNAs in CEM-SS and MT-4 T-cells. The effects of miRNA mimic transfections combined with Enoxacin treatment on HIV-1 replication were assessed. Finally, we employed an in vitro DICER1 cleavage assay to study the effects of Enoxacin on a pro-HIV-1 miRNA hsa-miR-132 processing.
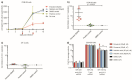
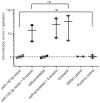
Results: We established that Enoxacin, but not the structurally similar compound nalidixic acid, exhibits strong anti-HIV-1 effects in the T-cell line CEM-SS, but not MT-4. We provide experimental data that this effect of Enoxacin is partly attributed to the specific downregulation of mature hsa-miR-132-3p, but not other tested pro- or anti-HIV-1 miRNAs, which is likely due to affecting DICER1 processing.
Conclusions: Our findings show an anti-retroviral activity of Enoxacin at least in part by downregulation of hsa-miR-132-3p, which may be relevant for future antiviral therapeutic applications by modulation of the RNA interference pathway.
Keywords: DICER1 processing; Enoxacin; HIV-1; RNA interference; fluoroquinolones; hsa-miR-132; miRNA.
Conflict of interest statement
K.J.M. has received travel grants and honoraria from Gilead Sciences, Roche Diagnostics, GlaxoSmithKline, Merck Sharp & Dohme, Bristol-Myers Squibb, ViiV and Abbott; and the University of Zurich received research grants from Gilead Science, Roche, and Merck Sharp & Dohme for studies that Dr. Metzner serves as principal investigator, and advisory board honoraria from Gilead Sciences. All other authors declare no conflicts of interest.
Figures



References
-
- UNAIDS UN Joint Programme on HIV/AIDS (UNAIDS), Global AIDS Update—2016. June 2016. [(accessed on 11 November 2024)]. Available online: https://www.refworld.org/reference/annualreport/unaids/2016/en/110274.
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

