Discovery of three novel neutralizing antibody epitopes on the human astrovirus capsid spike and mechanistic insights into virus neutralization
- PMID: 39846739
- PMCID: PMC11852706
- DOI: 10.1128/jvi.01619-24
Discovery of three novel neutralizing antibody epitopes on the human astrovirus capsid spike and mechanistic insights into virus neutralization
Abstract
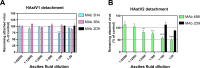
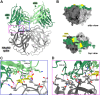
Human astroviruses (HAstVs) are a leading cause of viral childhood diarrhea that infects nearly every individual during their lifetime. Although human astroviruses are highly prevalent, no approved vaccine currently exists. Antibody responses appear to play an important role in protection from HAstV infection; however, knowledge about the neutralizing epitope landscape is lacking, as only three neutralizing antibody epitopes have previously been determined. Here, we structurally define the epitopes of three uncharacterized HAstV-neutralizing monoclonal antibodies: antibody 4B6 with X-ray crystallography to 2.67 Å, and antibodies 3H4 and 3B4 simultaneously with single-particle cryogenic-electron microscopy to 3.33 Å. We assess the epitope locations relative to conserved regions on the capsid spike and find that while antibodies 4B6 and 3B4 target the upper variable loop regions of the HAstV spike protein, antibody 3H4 targets a novel region near the base of the spike that is more conserved. Additionally, we found that all three antibodies bind with high affinity, and they compete with receptor FcRn binding to the capsid spike. These studies inform which regions of the HAstV capsid can be targeted by monoclonal antibody therapies and could aid in rational vaccine design.IMPORTANCEHuman astroviruses (HAstVs) infect nearly every child in the world, causing diarrhea, vomiting, and fever. Despite the prevalence of human astroviruses, little is known about how antibodies block virus infection. Here, we determined high-resolution structures of the astrovirus capsid protein in a complex with three virus-neutralizing antibodies. The antibodies bind distinct sites on the capsid spike domain. The antibodies block virus attachment to human cells and prevent capsid spike interaction with the human neonatal Fc receptor. These findings support the use of the human astrovirus capsid spike as an antigen in a vaccine to prevent astrovirus disease.
Keywords: X-ray crystallography; capsid; cryo-EM; human astrovirus; monoclonal antibodies; neutralization; protein structure-function; surface antigens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
Discovery of three novel neutralizing antibody epitopes on the human astrovirus capsid spike and mechanistic insights into virus neutralization.bioRxiv [Preprint]. 2024 Sep 14:2024.09.14.613010. doi: 10.1101/2024.09.14.613010. bioRxiv. 2024. Update in: J Virol. 2025 Feb 25;99(2):e0161924. doi: 10.1128/jvi.01619-24. PMID: 39314284 Free PMC article. Updated. Preprint.
References
-
- Ghosh S, Kumar M, Santiana M, Mishra A, Zhang M, Labayo H, Chibly AM, Nakamura H, Tanaka T, Henderson W, Lewis E, Voss O, Su Y, Belkaid Y, Chiorini JA, Hoffman MP, Altan-Bonnet N. 2022. Enteric viruses replicate in salivary glands and infect through saliva. Nature New Biol 607:345–350. doi:10.1038/s41586-022-04895-8 - DOI - PMC - PubMed

