Gut phages and their interactions with bacterial and mammalian hosts
- PMID: 39846747
- PMCID: PMC11844821
- DOI: 10.1128/jb.00428-24
Gut phages and their interactions with bacterial and mammalian hosts
Abstract
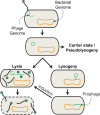
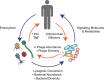
The mammalian gut microbiome is a dense and diverse community of microorganisms that reside in the distal gastrointestinal tract. In recent decades, the bacterial members of the gut microbiome have been the subject of intense research. Less well studied is the large community of bacteriophages that reside in the gut, which number in the billions of viral particles per gram of feces, and consist of considerable unknown viral "dark matter." This community of gut-residing bacteriophages, called the gut "phageome," plays a central role in the gut microbiome through predation and transformation of native gut bacteria, and through interactions with their mammalian hosts. In this review, we will summarize what is known about the composition and origins of the gut phageome, as well as its role in microbiome homeostasis and host health. Furthermore, we will outline the interactions of gut phages with their bacterial and mammalian hosts, and plot a course for the mechanistic study of these systems.
Keywords: microbiome; phages.
Conflict of interest statement
A.J.M. is a co-founder of Profluent Bio. The other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

