Reduced autoimmunity associated with deletion of host CD73
- PMID: 39846845
- PMCID: PMC11841978
- DOI: 10.1093/immhor/vlae004
Reduced autoimmunity associated with deletion of host CD73
Abstract
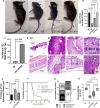
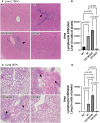
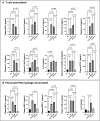
CD73 is ubiquitously expressed and regulates critical functions across multiple organ systems. The sequential actions of CD39 and CD73 accomplish the conversion of adenosine triphosphate to adenosine and shift the adenosine triphosphate-driven proinflammatory immune cell milieu toward an anti-inflammatory state. This immunological switch is a major mechanism by which regulatory T (Treg) cells control inflammation. Foxp3 engages in Treg development and function. Foxp3 mutations result in the scurfy (SF) mouse phenotype and a rapidly lethal lymphoproliferative syndrome. We generated double knockout (KO) mouse (CD73KOSF) by breeding heterozygous Foxp3sf/J females to CD73KO male mice to remove host CD73. We initially aimed to use these mice to identify a specific probiotic-CD73 effect, previously shown for Limosilactobacillus reuteri DSM 17938. We expected CD73 deletion to enhance the severity of autoimmunity in SF mice. However, we unexpectedly observed that KO of host CD73 in SF mice clinically reduced the severity of autoimmunity including reduced ear thickness, increased ear size, and less deformed ears, along with less dry and brittle skin. KO of CD73 in SF mice significantly reduced the numbers of CD4+ and CD8+T cells in spleen and blood. We identified that KO of CD73 in SF mice reduced the numbers of T cells in the thymus compared with those in SF mice, indicating that the milder clinical phenotype may be due to reduced central and peripheral lymphoproliferation. These new findings suggest targeting CD73 could improve T cell-mediated dermatitis, one of the most common symptoms in Treg deficiency-associated primary immune deficiencies.
Keywords: IPEX syndrome; dermatitis; primary immune deficiency; regulatory T cell; scurfy; thymocytes.
© The Author(s) 2025. Published by Oxford University Press on behalf of The American Association of Immunologists.
Conflict of interest statement
All authors have no financial conflict of interest to disclose.
Figures



References
-
- Dikiy S, Rudensky AY. Principles of regulatory T cell function. Immunity. 2023;56:240–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

